+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20756 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | SAM-bound SAM-IV riboswitch | ||||||||||||
 Map data Map data | SAM-bound SAM-IV riboswitch | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | SAM-IV riboswitch / Cryo-EM / Small RNA / RNA | ||||||||||||
| Biological species |   Mycobacterium sp. MCS (bacteria) Mycobacterium sp. MCS (bacteria) | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.1 Å | ||||||||||||
 Authors Authors | Zhang K / Li S | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2019 Journal: Nat Commun / Year: 2019Title: Cryo-EM structure of a 40 kDa SAM-IV riboswitch RNA at 3.7 Å resolution. Authors: Kaiming Zhang / Shanshan Li / Kalli Kappel / Grigore Pintilie / Zhaoming Su / Tung-Chung Mou / Michael F Schmid / Rhiju Das / Wah Chiu /  Abstract: Specimens below 50 kDa have generally been considered too small to be analyzed by single-particle cryo-electron microscopy (cryo-EM). The high flexibility of pure RNAs makes it difficult to obtain ...Specimens below 50 kDa have generally been considered too small to be analyzed by single-particle cryo-electron microscopy (cryo-EM). The high flexibility of pure RNAs makes it difficult to obtain high-resolution structures by cryo-EM. In bacteria, riboswitches regulate sulfur metabolism through binding to the S-adenosylmethionine (SAM) ligand and offer compelling targets for new antibiotics. SAM-I, SAM-I/IV, and SAM-IV are the three most commonly found SAM riboswitches, but the structure of SAM-IV is still unknown. Here, we report the structures of apo and SAM-bound SAM-IV riboswitches (119-nt, ~40 kDa) to 3.7 Å and 4.1 Å resolution, respectively, using cryo-EM. The structures illustrate homologies in the ligand-binding core but distinct peripheral tertiary contacts in SAM-IV compared to SAM-I and SAM-I/IV. Our results demonstrate the feasibility of resolving small RNAs with enough detail to enable detection of their ligand-binding pockets and suggest that cryo-EM could play a role in structure-assisted drug design for RNA. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20756.map.gz emd_20756.map.gz | 17 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20756-v30.xml emd-20756-v30.xml emd-20756.xml emd-20756.xml | 13.9 KB 13.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_20756.png emd_20756.png | 114.2 KB | ||
| Filedesc metadata |  emd-20756.cif.gz emd-20756.cif.gz | 4.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20756 http://ftp.pdbj.org/pub/emdb/structures/EMD-20756 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20756 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20756 | HTTPS FTP |
-Related structure data
| Related structure data |  6uetMC  6uesC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_20756.map.gz / Format: CCP4 / Size: 18.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20756.map.gz / Format: CCP4 / Size: 18.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | SAM-bound SAM-IV riboswitch | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
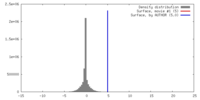
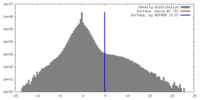
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : SAM-bound SAM-IV riboswitch
| Entire | Name: SAM-bound SAM-IV riboswitch |
|---|---|
| Components |
|
-Supramolecule #1: SAM-bound SAM-IV riboswitch
| Supramolecule | Name: SAM-bound SAM-IV riboswitch / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 40 KDa |
-Supramolecule #2: SAM-bound SAM-IV riboswitch
| Supramolecule | Name: SAM-bound SAM-IV riboswitch / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: RNA (119-MER)
| Macromolecule | Name: RNA (119-MER) / type: rna / ID: 1 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Mycobacterium sp. MCS (bacteria) Mycobacterium sp. MCS (bacteria) |
| Molecular weight | Theoretical: 38.522906 KDa |
| Sequence | String: GGUCAUGAGU GCCAGCGUCA AGCCCCGGCU UGCUGGCCGG CAACCCUCCA ACCGCGGUGG GGUGCCCCGG GUGAUGACCA GGUUGAGUA GCCGUGACGG CUACGCGGCA AGCGCGGGUC GENBANK: GENBANK: CP000384.1 |
-Macromolecule #2: S-ADENOSYLMETHIONINE
| Macromolecule | Name: S-ADENOSYLMETHIONINE / type: ligand / ID: 2 / Number of copies: 1 / Formula: SAM |
|---|---|
| Molecular weight | Theoretical: 398.437 Da |
| Chemical component information |  ChemComp-SAM: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.2 mg/mL |
|---|---|
| Buffer | pH: 7.2 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK IV |
| Details | SAM-bound SAM-IV riboswitch |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Frames/image: 1-30 / Number grids imaged: 2 / Number real images: 6030 / Average exposure time: 6.0 sec. / Average electron dose: 7.6 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: BACKBONE TRACE |
|---|---|
| Output model |  PDB-6uet: |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)