+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20616 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | EM structure of MPEG-1(w.t.) soluble pre-pore | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | MPEG-1/Perforin-2 / MACPF / macrophage / pore forming proteins / immunity / IMMUNE SYSTEM | |||||||||
| Function / homology |  Function and homology information Function and homology informationdendritic cell antigen processing and presentation / antigen processing and presentation of exogenous peptide antigen / phagolysosome membrane / antibacterial innate immune response / wide pore channel activity / antigen processing and presentation of exogenous peptide antigen via MHC class I / phagocytic vesicle / phagocytic vesicle membrane / cytoplasmic vesicle / defense response to Gram-negative bacterium ...dendritic cell antigen processing and presentation / antigen processing and presentation of exogenous peptide antigen / phagolysosome membrane / antibacterial innate immune response / wide pore channel activity / antigen processing and presentation of exogenous peptide antigen via MHC class I / phagocytic vesicle / phagocytic vesicle membrane / cytoplasmic vesicle / defense response to Gram-negative bacterium / adaptive immune response / defense response to bacterium / defense response to Gram-positive bacterium / extracellular region Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
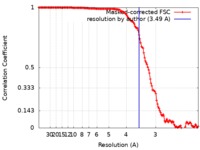
| Method | single particle reconstruction / cryo EM / Resolution: 3.49 Å | |||||||||
 Authors Authors | Pang SS / Bayly-Jones C | |||||||||
| Funding support |  Australia, 1 items Australia, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2019 Journal: Nat Commun / Year: 2019Title: The cryo-EM structure of the acid activatable pore-forming immune effector Macrophage-expressed gene 1. Authors: Siew Siew Pang / Charles Bayly-Jones / Mazdak Radjainia / Bradley A Spicer / Ruby H P Law / Adrian W Hodel / Edward S Parsons / Susan M Ekkel / Paul J Conroy / Georg Ramm / Hariprasad ...Authors: Siew Siew Pang / Charles Bayly-Jones / Mazdak Radjainia / Bradley A Spicer / Ruby H P Law / Adrian W Hodel / Edward S Parsons / Susan M Ekkel / Paul J Conroy / Georg Ramm / Hariprasad Venugopal / Phillip I Bird / Bart W Hoogenboom / Ilia Voskoboinik / Yann Gambin / Emma Sierecki / Michelle A Dunstone / James C Whisstock /    Abstract: Macrophage-expressed gene 1 (MPEG1/Perforin-2) is a perforin-like protein that functions within the phagolysosome to damage engulfed microbes. MPEG1 is thought to form pores in target membranes, ...Macrophage-expressed gene 1 (MPEG1/Perforin-2) is a perforin-like protein that functions within the phagolysosome to damage engulfed microbes. MPEG1 is thought to form pores in target membranes, however, its mode of action remains unknown. We use cryo-Electron Microscopy (cryo-EM) to determine the 2.4 Å structure of a hexadecameric assembly of MPEG1 that displays the expected features of a soluble prepore complex. We further discover that MPEG1 prepore-like assemblies can be induced to perforate membranes through acidification, such as would occur within maturing phagolysosomes. We next solve the 3.6 Å cryo-EM structure of MPEG1 in complex with liposomes. These data reveal that a multi-vesicular body of 12 kDa (MVB12)-associated β-prism (MABP) domain binds membranes such that the pore-forming machinery of MPEG1 is oriented away from the bound membrane. This unexpected mechanism of membrane interaction suggests that MPEG1 remains bound to the phagolysosome membrane while simultaneously forming pores in engulfed bacterial targets. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20616.map.gz emd_20616.map.gz | 229.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20616-v30.xml emd-20616-v30.xml emd-20616.xml emd-20616.xml | 18.2 KB 18.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_20616_fsc.xml emd_20616_fsc.xml | 13.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_20616.png emd_20616.png | 110.5 KB | ||
| Masks |  emd_20616_msk_1.map emd_20616_msk_1.map | 244.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-20616.cif.gz emd-20616.cif.gz | 6.1 KB | ||
| Others |  emd_20616_additional.map.gz emd_20616_additional.map.gz emd_20616_half_map_1.map.gz emd_20616_half_map_1.map.gz emd_20616_half_map_2.map.gz emd_20616_half_map_2.map.gz | 117.9 MB 225 MB 224.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20616 http://ftp.pdbj.org/pub/emdb/structures/EMD-20616 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20616 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20616 | HTTPS FTP |
-Related structure data
| Related structure data |  6u23MC  6u2jC  6u2kC  6u2lC  6u2wC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_20616.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20616.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
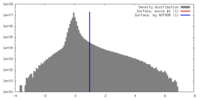
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_20616_msk_1.map emd_20616_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
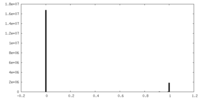
| Density Histograms |
-Additional map: unsharpened map
| File | emd_20616_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
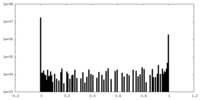
| Density Histograms |
-Half map: #2
| File | emd_20616_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
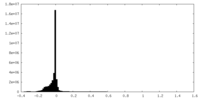
| Density Histograms |
-Half map: #1
| File | emd_20616_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Macrophage-Expressed Gene 1 (MPEG-1) 16-mer soluble pre-pore complex
| Entire | Name: Macrophage-Expressed Gene 1 (MPEG-1) 16-mer soluble pre-pore complex |
|---|---|
| Components |
|
-Supramolecule #1: Macrophage-Expressed Gene 1 (MPEG-1) 16-mer soluble pre-pore complex
| Supramolecule | Name: Macrophage-Expressed Gene 1 (MPEG-1) 16-mer soluble pre-pore complex type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 2.3 MDa |
-Macromolecule #1: Macrophage-expressed gene 1 protein
| Macromolecule | Name: Macrophage-expressed gene 1 protein / type: protein_or_peptide / ID: 1 / Number of copies: 16 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 71.110125 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: KSGKPSGEMD EVGVQKCKNA LKLPVLEVLP GGGWDNLRNV DMGRVMELTY SNCRTTEDGQ YIIPDEIFTI PQKQSNLEMN SEILESWAN YQSSTSYSIN TELSLFSKVN GKFSTEFQRM KTLQVKDQAI TTRVQVRNLV YTVKINPTLE LSSGFRKELL D ISDRLENN ...String: KSGKPSGEMD EVGVQKCKNA LKLPVLEVLP GGGWDNLRNV DMGRVMELTY SNCRTTEDGQ YIIPDEIFTI PQKQSNLEMN SEILESWAN YQSSTSYSIN TELSLFSKVN GKFSTEFQRM KTLQVKDQAI TTRVQVRNLV YTVKINPTLE LSSGFRKELL D ISDRLENN QTRMATYLAE LLVLNYGTHV TTSVDAGAAL IQEDHLRASF LQDSQSSRSA VTASAGLAFQ NTVNFKFEEN YT SQNVLTK SYLSNRTNSR VQSIGGVPFY PGITLQAWQQ GITNHLVAID RSGLPLHFFI NPNMLPDLPG PLVKKVSKTV ETA VKRYYT FNTYPGCTDL NSPNFNFQAN TDDGSCEGKM TNFSFGGVYQ ECTQLSGNRD VLLCQKLEQK NPLTGDFSCP SGYS PVHLL SQIHEEGYNH LECHRKCTLL VFCKTVCEDV FQVAKAEFRA FWCVASSQVP ENSGLLFGGL FSSKSINPMT NAQSC PAGY FPLRLFENLK VCVSQDYELG SRFAVPFGGF FSCTVGNPLV DPAISRDLGA PSLKKCPGGF SQHPALISDG CQVSYC VKS GLFTGGSLPP ARLPPFTRPP LMSQAATNTV IVTNSENARS WIKDSQTHQW RLGEPIELRR AMNVIHGDGG GLSHHHH HH UniProtKB: Macrophage-expressed gene 1 protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.2 Component:
| |||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV | |||||||||
| Details | This sample was monodisperse |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Average electron dose: 45.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-6u23: |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)