+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20583 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | ExbB-ExbD complex in MSP1E3D1 nanodisc | |||||||||
 Map data Map data | ExbB-ExbD complex in MSP1E3D1 nanodisc | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | molecular motor / Ton system / membrane protein / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationferrichrome import into cell / energy transducer activity / bacteriocin transport / cobalamin transport / intracellular monoatomic cation homeostasis / transmembrane transporter complex / protein import / plasma membrane protein complex / transmembrane transporter activity / cell outer membrane ...ferrichrome import into cell / energy transducer activity / bacteriocin transport / cobalamin transport / intracellular monoatomic cation homeostasis / transmembrane transporter complex / protein import / plasma membrane protein complex / transmembrane transporter activity / cell outer membrane / protein transport / intracellular iron ion homeostasis / protein stabilization / protein homodimerization activity / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |   | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Celia H / Botos I | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Commun Biol / Year: 2019 Journal: Commun Biol / Year: 2019Title: Cryo-EM structure of the bacterial Ton motor subcomplex ExbB-ExbD provides information on structure and stoichiometry. Authors: Herve Celia / Istvan Botos / Xiaodan Ni / Tara Fox / Natalia De Val / Roland Lloubes / Jiansen Jiang / Susan K Buchanan /   Abstract: The TonB-ExbB-ExbD molecular motor harnesses the proton motive force across the bacterial inner membrane to couple energy to transporters at the outer membrane, facilitating uptake of essential ...The TonB-ExbB-ExbD molecular motor harnesses the proton motive force across the bacterial inner membrane to couple energy to transporters at the outer membrane, facilitating uptake of essential nutrients such as iron and cobalamine. TonB physically interacts with the nutrient-loaded transporter to exert a force that opens an import pathway across the outer membrane. Until recently, no high-resolution structural information was available for this unique molecular motor. We published the first crystal structure of ExbB-ExbD in 2016 and showed that five copies of ExbB are arranged as a pentamer around a single copy of ExbD. However, our spectroscopic experiments clearly indicated that two copies of ExbD are present in the complex. To resolve this ambiguity, we used single-particle cryo-electron microscopy to show that the ExbB pentamer encloses a dimer of ExbD in its transmembrane pore, and not a monomer as previously reported. The revised stoichiometry has implications for motor function. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20583.map.gz emd_20583.map.gz | 1.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20583-v30.xml emd-20583-v30.xml emd-20583.xml emd-20583.xml | 14.7 KB 14.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_20583.png emd_20583.png | 123 KB | ||
| Filedesc metadata |  emd-20583.cif.gz emd-20583.cif.gz | 6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20583 http://ftp.pdbj.org/pub/emdb/structures/EMD-20583 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20583 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20583 | HTTPS FTP |
-Related structure data
| Related structure data |  6tyiMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_20583.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20583.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | ExbB-ExbD complex in MSP1E3D1 nanodisc | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
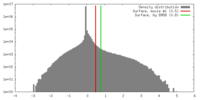
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Ton subcomplex ExbB-ExbD reconstituted in lipid nanodisc
| Entire | Name: Ton subcomplex ExbB-ExbD reconstituted in lipid nanodisc |
|---|---|
| Components |
|
-Supramolecule #1: Ton subcomplex ExbB-ExbD reconstituted in lipid nanodisc
| Supramolecule | Name: Ton subcomplex ExbB-ExbD reconstituted in lipid nanodisc type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Biopolymer transport protein ExbB
| Macromolecule | Name: Biopolymer transport protein ExbB / type: protein_or_peptide / ID: 1 / Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 26.312322 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGNNLMQTDL SVWGMYQHAD IVVKCVMIGL ILASVVTWAI FFSKSVEFFN QKRRLKREQQ LLAEARSLNQ ANDIAADFGS KSLSLHLLN EAQNELELSE GSDDNEGIKE RTSFRLERRV AAVGRQMGRG NGYLATIGAI SPFVGLFGTV WGIMNSFIGI A QTQTTNLA ...String: MGNNLMQTDL SVWGMYQHAD IVVKCVMIGL ILASVVTWAI FFSKSVEFFN QKRRLKREQQ LLAEARSLNQ ANDIAADFGS KSLSLHLLN EAQNELELSE GSDDNEGIKE RTSFRLERRV AAVGRQMGRG NGYLATIGAI SPFVGLFGTV WGIMNSFIGI A QTQTTNLA VVAPGIAEAL LATAIGLVAA IPAVVIYNVF ARQIGGFKAM LGDVAAQVLL LQSRDLDLEA SAAAHPVRVA QK LRAG UniProtKB: Biopolymer transport protein ExbB |
-Macromolecule #2: Biopolymer transport protein ExbD
| Macromolecule | Name: Biopolymer transport protein ExbD / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 18.161674 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAMHLNENLD DNGEMHDINV TPFIDVMLVL LIIFMVAAPL ATVDVKVNLP ASTSTPQPRP EKPVYLSVKA DNSMFIGNDP VTDETMITA LNALTEGKKD TTIFFRADKT VDYETLMKVM DTLHQAGYLK IGLVGEETAK AKENLYFQGN AGSGHHHHHH H HHH UniProtKB: Biopolymer transport protein ExbD |
-Macromolecule #3: (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-...
| Macromolecule | Name: (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE type: ligand / ID: 3 / Number of copies: 1 / Formula: PGT |
|---|---|
| Molecular weight | Theoretical: 751.023 Da |
| Chemical component information |  ChemComp-PGT: |
-Macromolecule #4: (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY...
| Macromolecule | Name: (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE type: ligand / ID: 4 / Number of copies: 3 / Formula: PEV |
|---|---|
| Molecular weight | Theoretical: 720.012 Da |
| Chemical component information |  ChemComp-PEV: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 1.6 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)