+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20563 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | TRIM5alpha dimer in complex with HIV-1 capsid tubes | |||||||||||||||
 Map data Map data | TRIM5alpha dimer | |||||||||||||||
 Sample Sample |
| |||||||||||||||
| Biological species |   Human immunodeficiency virus 1 Human immunodeficiency virus 1 | |||||||||||||||
| Method | electron tomography / cryo EM | |||||||||||||||
 Authors Authors | Skorupka KA / Pornillos O / Ganser-Pornillos BK | |||||||||||||||
| Funding support |  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2019 Journal: Sci Adv / Year: 2019Title: Hierarchical assembly governs TRIM5α recognition of HIV-1 and retroviral capsids. Authors: Katarzyna A Skorupka / Marcin D Roganowicz / Devin E Christensen / Yueping Wan / Owen Pornillos / Barbie K Ganser-Pornillos /  Abstract: TRIM5α is a restriction factor that senses incoming retrovirus cores through an unprecedented mechanism of nonself recognition. TRIM5α assembles a hexagonal lattice that avidly binds the capsid ...TRIM5α is a restriction factor that senses incoming retrovirus cores through an unprecedented mechanism of nonself recognition. TRIM5α assembles a hexagonal lattice that avidly binds the capsid shell, which surrounds and protects the virus core. The extent to which the TRIM lattice can cover the capsid and how TRIM5α directly contacts the capsid surface have not been established. Here, we apply cryo-electron tomography and subtomogram averaging to determine structures of TRIM5α bound to recombinant HIV-1 capsid assemblies. Our data support a mechanism of hierarchical assembly, in which a limited number of basal interaction modes are successively organized in increasingly higher-order structures that culminate in a TRIM5α cage surrounding a retroviral capsid. We further propose that cage formation explains the mechanism of restriction and provides the structural context that links capsid recognition to ubiquitin-dependent processes that disable the retrovirus. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20563.map.gz emd_20563.map.gz | 6.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20563-v30.xml emd-20563-v30.xml emd-20563.xml emd-20563.xml | 12.8 KB 12.8 KB | Display Display |  EMDB header EMDB header |
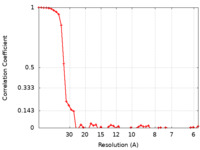
| FSC (resolution estimation) |  emd_20563_fsc.xml emd_20563_fsc.xml | 5.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_20563.png emd_20563.png | 94.1 KB | ||
| Others |  emd_20563_half_map_1.map.gz emd_20563_half_map_1.map.gz emd_20563_half_map_2.map.gz emd_20563_half_map_2.map.gz | 6.9 MB 6.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20563 http://ftp.pdbj.org/pub/emdb/structures/EMD-20563 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20563 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20563 | HTTPS FTP |
-Validation report
| Summary document |  emd_20563_validation.pdf.gz emd_20563_validation.pdf.gz | 78 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_20563_full_validation.pdf.gz emd_20563_full_validation.pdf.gz | 77.1 KB | Display | |
| Data in XML |  emd_20563_validation.xml.gz emd_20563_validation.xml.gz | 500 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20563 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20563 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20563 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20563 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_20563.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20563.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | TRIM5alpha dimer | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.92 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
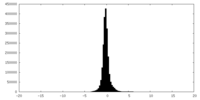
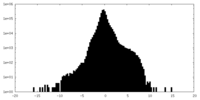
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: TRIM5alpha dimer odd half-set
| File | emd_20563_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | TRIM5alpha dimer odd half-set | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: TRIM5alpha dimer even half-set
| File | emd_20563_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | TRIM5alpha dimer even half-set | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Human immunodeficiency virus 1
| Entire | Name:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
|---|---|
| Components |
|
-Supramolecule #1: Human immunodeficiency virus 1
| Supramolecule | Name: Human immunodeficiency virus 1 / type: virus / ID: 1 / Parent: 0 / NCBI-ID: 11676 / Sci species name: Human immunodeficiency virus 1 / Virus type: VIRUS-LIKE PARTICLE / Virus isolate: OTHER / Virus enveloped: No / Virus empty: Yes |
|---|---|
| Host system | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | electron tomography |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
| Sectioning | Other: NO SECTIONING |
| Fiducial marker | Manufacturer: Nanoprobes / Diameter: 10 nm |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Average electron dose: 1.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)