[English] 日本語
 Yorodumi
Yorodumi- EMDB-20503: Subtomogram average of a purified Leptospira biflexa -fcpB mutant... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20503 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Subtomogram average of a purified Leptospira biflexa -fcpB mutant flagellum | |||||||||
 Map data Map data | Map of the Leptospira biflexa purified -fcpB flagellum, determined through subtomogram averaging with emClarity. | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Leptospira biflexa serovar Patoc (bacteria) Leptospira biflexa serovar Patoc (bacteria) | |||||||||
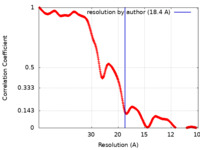
| Method | subtomogram averaging / cryo EM / Resolution: 18.4 Å | |||||||||
 Authors Authors | Brady MR / Gibson KH / Sindelar CV | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation | Journal: Front Cell Infect Microbiol / Year: 2018 Title: FcpB Is a Surface Filament Protein of the Endoflagellum Required for the Motility of the Spirochete . Authors: Elsio A Wunder / Leyla Slamti / David N Suwondo / Kimberley H Gibson / Zhiguo Shang / Charles V Sindelar / Felipe Trajtenberg / Alejandro Buschiazzo / Albert I Ko / Mathieu Picardeau /    Abstract: The spirochete endoflagellum is a unique motility apparatus among bacteria. Despite its critical importance for pathogenesis, the full composition of the flagellum remains to be determined. We have ...The spirochete endoflagellum is a unique motility apparatus among bacteria. Despite its critical importance for pathogenesis, the full composition of the flagellum remains to be determined. We have recently reported that FcpA is a novel flagellar protein and a major component of the sheath of the filament of the spirochete . By screening a library of random transposon mutants in the spirochete , we found a motility-deficient mutant harboring a disruption in a hypothetical gene of unknown function. Here, we show that this gene encodes a surface component of the endoflagellar filament and is required for typical hook- and spiral-shaped ends of the cell body, coiled structure of the endoflagella, and high velocity phenotype. We therefore named the gene for flagellar-coiling protein B. is conserved in all members of the genus, but not present in other organisms including other spirochetes. Complementation of the mutant restored the wild-type morphology and motility phenotypes. Immunoblotting with anti-FcpA and anti-FcpB antisera and cryo-electron microscopy of the filament indicated that FcpB assembled onto the surface of the sheath of the filament and mostly located on the outer (convex) side of the coiled filament. We provide evidence that FcpB, together with FcpA, are -specific novel components of the sheath of the filament, key determinants of the coiled and asymmetric structure of the endoflagella and are essential for high velocity. Defining the components of the endoflagella and their functions in these atypical bacteria should greatly enhance our understanding of the mechanisms by which these bacteria produce motility. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20503.map.gz emd_20503.map.gz | 18 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20503-v30.xml emd-20503-v30.xml emd-20503.xml emd-20503.xml | 17.1 KB 17.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_20503_fsc.xml emd_20503_fsc.xml | 33.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_20503.png emd_20503.png | 24.9 KB | ||
| Others |  emd_20503_half_map_1.map.gz emd_20503_half_map_1.map.gz emd_20503_half_map_2.map.gz emd_20503_half_map_2.map.gz | 18 MB 18 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20503 http://ftp.pdbj.org/pub/emdb/structures/EMD-20503 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20503 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20503 | HTTPS FTP |
-Validation report
| Summary document |  emd_20503_validation.pdf.gz emd_20503_validation.pdf.gz | 78.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_20503_full_validation.pdf.gz emd_20503_full_validation.pdf.gz | 77.3 KB | Display | |
| Data in XML |  emd_20503_validation.xml.gz emd_20503_validation.xml.gz | 496 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20503 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20503 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20503 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20503 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_20503.map.gz / Format: CCP4 / Size: 19.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20503.map.gz / Format: CCP4 / Size: 19.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map of the Leptospira biflexa purified -fcpB flagellum, determined through subtomogram averaging with emClarity. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.97 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
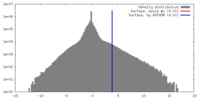
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: Half mask of the Leptospira biflexa purified -fcpB...
| File | emd_20503_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half mask of the Leptospira biflexa purified -fcpB flagellum, determined through subtomogram averaging with emClarity. | ||||||||||||
| Projections & Slices |
| ||||||||||||
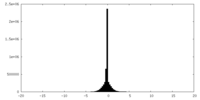
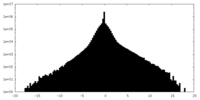
| Density Histograms |
-Half map: Half mask of the Leptospira biflexa purified -fcpB...
| File | emd_20503_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half mask of the Leptospira biflexa purified -fcpB flagellum, determined through subtomogram averaging with emClarity. | ||||||||||||
| Projections & Slices |
| ||||||||||||
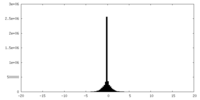
| Density Histograms |
- Sample components
Sample components
-Entire : Flagellum
| Entire | Name: Flagellum |
|---|---|
| Components |
|
-Supramolecule #1: Flagellum
| Supramolecule | Name: Flagellum / type: complex / ID: 1 / Parent: 0 Details: Purified periplasmic flagella from Leptospira biflexa serovar Patoc -fcpB mutant |
|---|---|
| Source (natural) | Organism:  Leptospira biflexa serovar Patoc (bacteria) / Location in cell: Periplasmic space Leptospira biflexa serovar Patoc (bacteria) / Location in cell: Periplasmic space |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 6.8 / Component - Formula: H2O |
|---|---|
| Grid | Model: C-flat-1.2/1.3 / Material: COPPER / Mesh: 200 / Support film - #0 - Film type ID: 1 / Support film - #0 - Material: CARBON / Support film - #0 - topology: CONTINUOUS / Support film - #0 - Film thickness: 3.0 nm / Support film - #1 - Film type ID: 2 / Support film - #1 - Material: CARBON / Support film - #1 - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE Details: The sample was glow-discharged with ArO2 for 6 seconds. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 291 K / Instrument: FEI VITROBOT MARK III / Details: Blot time of 6 seconds, blot offset -1mm. |
| Details | Flagella were purified from Leptospira biflexa serovar Patoc according to the method described from Wunder et al. 2016 (Molecular Microbiology). After the final centrifugation, the flagella were resuspended in 0.5 mL H2O, and approximately 0.2% sodium azide was added as a preservative. Flagella were stored at 4 degrees C. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Average exposure time: 1.5 sec. / Average electron dose: 8.35 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 1.2 mm / Nominal defocus min: 2.5 µm / Nominal magnification: 14500 |
| Sample stage | Specimen holder model: GATAN 626 SINGLE TILT LIQUID NITROGEN CRYO TRANSFER HOLDER Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)