+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20447 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of AdnAB-AMPPNP-DNA complex | |||||||||
 Map data Map data | AdnAB-AMPPNP-DNA complex | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | DNA / DNA BINDING PROTEIN / DNA BINDING PROTEIN-DNA complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationDNA helicase complex / recombinational repair / exonuclease activity / DNA 3'-5' helicase / DNA helicase activity / isomerase activity / DNA helicase / forked DNA-dependent helicase activity / single-stranded 3'-5' DNA helicase activity / four-way junction helicase activity ...DNA helicase complex / recombinational repair / exonuclease activity / DNA 3'-5' helicase / DNA helicase activity / isomerase activity / DNA helicase / forked DNA-dependent helicase activity / single-stranded 3'-5' DNA helicase activity / four-way junction helicase activity / double-stranded DNA helicase activity / hydrolase activity / DNA repair / DNA binding / ATP binding / cytosol Similarity search - Function | |||||||||
| Biological species |  Mycolicibacterium smegmatis (bacteria) / Mycolicibacterium smegmatis (bacteria) /   Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155) (bacteria) / Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155) (bacteria) /  Mycobacterium smegmatis (bacteria) Mycobacterium smegmatis (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.5 Å | |||||||||
 Authors Authors | Jia N / Unciuleac M / Shuman S / Patel DJ | |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2019 Journal: Proc Natl Acad Sci U S A / Year: 2019Title: Structures and single-molecule analysis of bacterial motor nuclease AdnAB illuminate the mechanism of DNA double-strand break resection. Authors: Ning Jia / Mihaela C Unciuleac / Chaoyou Xue / Eric C Greene / Dinshaw J Patel / Stewart Shuman /  Abstract: Mycobacterial AdnAB is a heterodimeric helicase-nuclease that initiates homologous recombination by resecting DNA double-strand breaks (DSBs). The AdnA and AdnB subunits are each composed of an N- ...Mycobacterial AdnAB is a heterodimeric helicase-nuclease that initiates homologous recombination by resecting DNA double-strand breaks (DSBs). The AdnA and AdnB subunits are each composed of an N-terminal motor domain and a C-terminal nuclease domain. Here we report cryoelectron microscopy (cryo-EM) structures of AdnAB in three functional states: in the absence of DNA and in complex with forked duplex DNAs before and after cleavage of the 5' single-strand DNA (ssDNA) tail by the AdnA nuclease. The structures reveal the path of the 5' ssDNA through the AdnA nuclease domain and the mechanism of 5' strand cleavage; the path of the 3' tracking strand through the AdnB motor and the DNA contacts that couple ATP hydrolysis to mechanical work; the position of the AdnA iron-sulfur cluster subdomain at the Y junction and its likely role in maintaining the split trajectories of the unwound 5' and 3' strands. Single-molecule DNA curtain analysis of DSB resection reveals that AdnAB is highly processive but prone to spontaneous pausing at random sites on duplex DNA. A striking property of AdnAB is that the velocity of DSB resection slows after the enzyme experiences a spontaneous pause. Our results highlight shared as well as distinctive properties of AdnAB vis-à-vis the RecBCD and AddAB clades of bacterial DSB-resecting motor nucleases. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20447.map.gz emd_20447.map.gz | 6.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20447-v30.xml emd-20447-v30.xml emd-20447.xml emd-20447.xml | 15.6 KB 15.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_20447_fsc.xml emd_20447_fsc.xml | 10 KB | Display |  FSC data file FSC data file |
| Images |  emd_20447.png emd_20447.png | 70.8 KB | ||
| Filedesc metadata |  emd-20447.cif.gz emd-20447.cif.gz | 6.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20447 http://ftp.pdbj.org/pub/emdb/structures/EMD-20447 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20447 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20447 | HTTPS FTP |
-Related structure data
| Related structure data |  6ppuMC  6ppjC  6pprC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_20447.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20447.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | AdnAB-AMPPNP-DNA complex | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.8613 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
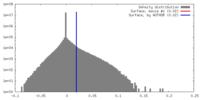
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : AdnAB-AMPPNP-DNA complex
| Entire | Name: AdnAB-AMPPNP-DNA complex |
|---|---|
| Components |
|
-Supramolecule #1: AdnAB-AMPPNP-DNA complex
| Supramolecule | Name: AdnAB-AMPPNP-DNA complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Molecular weight | Theoretical: 200 KDa |
-Supramolecule #2: UvrD/REP helicase
| Supramolecule | Name: UvrD/REP helicase / type: complex / ID: 2 / Parent: 1 |
|---|---|
| Source (natural) | Organism:  Mycolicibacterium smegmatis (bacteria) Mycolicibacterium smegmatis (bacteria) |
-Supramolecule #3: ATP-dependent DNA helicase (UvrD/REP)
| Supramolecule | Name: ATP-dependent DNA helicase (UvrD/REP) / type: complex / ID: 3 / Parent: 1 |
|---|---|
| Source (natural) | Organism:  Mycolicibacterium smegmatis (bacteria) Mycolicibacterium smegmatis (bacteria) |
-Supramolecule #4: DNA
| Supramolecule | Name: DNA / type: complex / ID: 4 / Parent: 1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: UvrD/REP helicase
| Macromolecule | Name: UvrD/REP helicase / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155) (bacteria) Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155) (bacteria)Strain: ATCC 700084 / mc(2)155 |
| Molecular weight | Theoretical: 118.128547 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTQVASPVVQ ARYSPVELSA ALGLFPPTDE QAAVIAAPPG PLVVIAGAGA GKTETMAARV VWLVANGFAT PSQVLGLTFT RKAAGQLLR RVRTRLARLA GAGLAPGSGA SDESATVSTY HAFAGTLLRE HGLLLPVEPD TRLLSETELW QLAYDVVCAH P GHLDTEKT ...String: MTQVASPVVQ ARYSPVELSA ALGLFPPTDE QAAVIAAPPG PLVVIAGAGA GKTETMAARV VWLVANGFAT PSQVLGLTFT RKAAGQLLR RVRTRLARLA GAGLAPGSGA SDESATVSTY HAFAGTLLRE HGLLLPVEPD TRLLSETELW QLAYDVVCAH P GHLDTEKT PAAVTAMVLR LSGALAEHLV DTDQLRDTHV ELERLVHTLP AGPYQRDRGP SQWLLRMLAT QTERTELVPL ID ALHQRMR AEKVMDFGMQ MAAAARLAAR FPQVGEQLRQ RFRVVLLDEY QDTGHAQRIA LSSLFGGGAD DGLALTAVGD PIQ SIYGWR GASATNLPRF TTDFPYSDGT PAPTLELRTS WRNPPSTLHV ANAVSEEARR RSVAVRALRP RPDAEPGTIR CALL NNVAA ERDWVADHLA RAYHGAIGRG EAAPTAAVLV RRNADAAPMA EALTARGVPV EVVGVAGLLA VPEVADLVAM LRLIA DPTA GSAVMRILTG PRWRFGARDI AALWRRAVEL DDRPKGELGT ADIVAQAAPD ADTACVADAI CDPGDAERYS PAGYER IVA LGRELTMLRA HLGHPLPELV AEVRRVLGLD AEARAARPVA AGWAGTENLD RFSDLVSDFA GHAGASVSAL LAYLDAA VE VENGLAPAEL TVSHDRVQIL TVHAAKGLEW QVVAVPHLSA RVFPSTTQAR TWLTDASDLP PLLRGDRATE SEIGVPVL D TSDIYDRKIL SDKISDHKKS LDQRRVDEER RLLYVAITRA EDTLLLSGHH WGATESKPRG PSEFLCELKT ILEEATAAG TPCGEIEHWA PDPAPGETNP LRDQVVEALW PPVASADDHV HRGAQLVAAA MAGEVSAEAD QEGWAADVDA LLAERERPPQ QEDTELPGQ LSVSTLVELS RDPKAALTRL RRRLPQRPDP HALLGTTFHE WVQRYFHAER LFDLDDLPGA VDSDSGRAVE E SLAELQDA FVKSPWAART PVEVEVPFDM VLGETVVRGR IDAVFAEPDG TTMVLDWKTG DPPETPEAKE HAAVQLAVYR LA WAAMRGC PPESVRAAFH YVRSGQTVIP ETLPGAEELV KLLAAAPTET AEEADRIT UniProtKB: DNA 3'-5' helicase |
-Macromolecule #2: ATP-dependent DNA helicase (UvrD/REP)
| Macromolecule | Name: ATP-dependent DNA helicase (UvrD/REP) / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO / EC number: DNA helicase |
|---|---|
| Source (natural) | Organism:  Mycobacterium smegmatis (bacteria) Mycobacterium smegmatis (bacteria) |
| Molecular weight | Theoretical: 76.049312 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) ...String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) GTVTVRLAAS THAEGTMIAD ALRRAHLVDG IPWSQMAVIV RSVPRVGTAL ARALTAAGV PVQDNGTDVP VGRQPAAAAL LTVLDVTATG HLDADSAVAL LTGPIGRVDP VTLRQLRRAL RRADGSQPPR D FGDLLVDA IEREPKGLSA EHARTLRRLR AVLTAARRSD ASGADPRYTL WQAWHASGLQ RRWLAASERG GSVGAQADRD LD AVTTLFD VADQYVNRTA GASLRGLVDH VTRLGAAVAR TEPETAAEAV AVLSVHGALA GEWDFVVIAG VQEGLWPNMI PRG GVLGTQ HLVDVLDGVA DMTDRTVSTR APLVAEERRL LMAAMGRART RVMITAVDS(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)PPLVAPR VLAPSALVGR LRAVVCAPDG AVDDDARACA AAQ LARLAA AGVPGADPSQ WHAMTSLTTE EPLWSEPGHV VTLSPSTLQM LTDCPLRWLL ERHGGDDGRD VRSTVGSLVH ALVS EPGKT ESQLVNELEK VWDDLPYDAK WYSDNELARH RAMLETFTRW REDTRRQLTE VATEIPVEGI VVEPGENTPG VRVRG RLDR LERDEAGRLV VVDLKTGKSP VTKDDAQNHA QLAMYQLAVA AGLLDDGDEP GGGKLVYLGK AGAAGATERE QDPLTP DKR AEWLETVGEA AAATAGPRFV ARVNNGCANC PVRSSCPAQA NGDRP UniProtKB: DNA helicase, DNA helicase |
-Macromolecule #3: DNA (29-MER)
| Macromolecule | Name: DNA (29-MER) / type: dna / ID: 3 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 21.477703 KDa |
| Sequence | String: (DT)(DT)(DT)(DT)(DT)(DT)(DT)(DC)(DT)(DA) (DA)(DT)(DG)(DC)(DG)(DA)(DG)(DC)(DA)(DC) (DT)(DG)(DC)(DT)(DA)(DT)(DT)(DC)(DC) (DC)(DT)(DA)(DG)(DC)(DA)(DG)(DT)(DG)(DC) (DT) (DC)(DG)(DC)(DA)(DT)(DT) ...String: (DT)(DT)(DT)(DT)(DT)(DT)(DT)(DC)(DT)(DA) (DA)(DT)(DG)(DC)(DG)(DA)(DG)(DC)(DA)(DC) (DT)(DG)(DC)(DT)(DA)(DT)(DT)(DC)(DC) (DC)(DT)(DA)(DG)(DC)(DA)(DG)(DT)(DG)(DC) (DT) (DC)(DG)(DC)(DA)(DT)(DT)(DA)(DG) (DA)(DT)(DT)(DT)(DT)(DG)(DT)(DT)(DT)(DT) (DT)(DT) (DT)(DA)(DG)(DC)(DG)(DG)(DT) (DT)(DT)(DT) |
-Macromolecule #4: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 4 / Number of copies: 1 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #5: IRON/SULFUR CLUSTER
| Macromolecule | Name: IRON/SULFUR CLUSTER / type: ligand / ID: 5 / Number of copies: 1 / Formula: SF4 |
|---|---|
| Molecular weight | Theoretical: 351.64 Da |
| Chemical component information |  ChemComp-FS1: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.5 mg/mL |
|---|---|
| Buffer | pH: 7.5 / Component - Formula: Tris / Details: 20 mM Tris-HCl, pH 7.5, 150 mM NaCl |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 2.16 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)