[English] 日本語
 Yorodumi
Yorodumi- EMDB-20163: The central pair apparatus focusing on the C1a-e-c supercomplex e... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20163 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
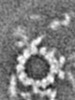
| Title | The central pair apparatus focusing on the C1a-e-c supercomplex extracted from the cryo-electron tomography and subtomographic average of isolated Chlamydomonas fap76-2 mutant axoneme | |||||||||||||||
 Map data Map data | The central pair apparatus focusing on the C1a-e-c supercomplex extracted from the cryo-electron tomography and subtomographic average of isolated Chlamydomonas fap76-2 mutant axoneme | |||||||||||||||
 Sample Sample |
| |||||||||||||||
| Biological species |  | |||||||||||||||
| Method | subtomogram averaging / cryo EM / Resolution: 26.0 Å | |||||||||||||||
 Authors Authors | Fu G / Nicastro D | |||||||||||||||
| Funding support |  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: J Cell Biol / Year: 2019 Journal: J Cell Biol / Year: 2019Title: Structural organization of the C1a-e-c supercomplex within the ciliary central apparatus. Authors: Gang Fu / Lei Zhao / Erin Dymek / Yuqing Hou / Kangkang Song / Nhan Phan / Zhiguo Shang / Elizabeth F Smith / George B Witman / Daniela Nicastro /  Abstract: Nearly all motile cilia contain a central apparatus (CA) composed of two connected singlet microtubules with attached projections that play crucial roles in regulating ciliary motility. Defects in CA ...Nearly all motile cilia contain a central apparatus (CA) composed of two connected singlet microtubules with attached projections that play crucial roles in regulating ciliary motility. Defects in CA assembly usually result in motility-impaired or paralyzed cilia, which in humans causes disease. Despite their importance, the protein composition and functions of the CA projections are largely unknown. Here, we integrated biochemical and genetic approaches with cryo-electron tomography to compare the CA of wild-type with CA mutants. We identified a large (>2 MD) complex, the C1a-e-c supercomplex, that requires the PF16 protein for assembly and contains the CA components FAP76, FAP81, FAP92, and FAP216. We localized these subunits within the supercomplex using nanogold labeling and show that loss of any one of them results in impaired ciliary motility. These data provide insight into the subunit organization and 3D structure of the CA, which is a prerequisite for understanding the molecular mechanisms by which the CA regulates ciliary beating. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20163.map.gz emd_20163.map.gz | 2.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20163-v30.xml emd-20163-v30.xml emd-20163.xml emd-20163.xml | 12.9 KB 12.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_20163.png emd_20163.png | 99.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20163 http://ftp.pdbj.org/pub/emdb/structures/EMD-20163 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20163 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20163 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_20163.map.gz / Format: CCP4 / Size: 3.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20163.map.gz / Format: CCP4 / Size: 3.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | The central pair apparatus focusing on the C1a-e-c supercomplex extracted from the cryo-electron tomography and subtomographic average of isolated Chlamydomonas fap76-2 mutant axoneme | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 5.523 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : The C1a-e-c supercomplex of central pair apparatus averaged from ...
| Entire | Name: The C1a-e-c supercomplex of central pair apparatus averaged from Chlamydomonas fap76-2 mutant cilia |
|---|---|
| Components |
|
-Supramolecule #1: The C1a-e-c supercomplex of central pair apparatus averaged from ...
| Supramolecule | Name: The C1a-e-c supercomplex of central pair apparatus averaged from Chlamydomonas fap76-2 mutant cilia type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Support film - Material: CARBON / Support film - topology: HOLEY / Details: unspecified |
| Vitrification | Cryogen name: ETHANE / Chamber temperature: 298 K / Instrument: HOMEMADE PLUNGER Details: back-side blotting with No.1 Whatman filter for 1.5-2.5 seconds before plunging. |
| Details | Freshly isolated and demembranated cilia from Chlamydomonas fap76-2 mutant cells |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Phase plate: VOLTA PHASE PLATE / Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3710 pixel / Digitization - Dimensions - Height: 3838 pixel / Digitization - Frames/image: 1-15 / Average exposure time: 6.0 sec. / Average electron dose: 1.57 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Calibrated defocus max: 0.5 µm / Calibrated defocus min: 0.2 µm / Calibrated magnification: 26000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Algorithm: BACK PROJECTION / Resolution.type: BY AUTHOR / Resolution: 26.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name:  IMOD / Number subtomograms used: 503 IMOD / Number subtomograms used: 503 |
|---|---|
| Extraction | Number tomograms: 10 / Number images used: 503 / Software - Name:  MATLAB MATLAB |
| Final angle assignment | Type: OTHER |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)