[English] 日本語
 Yorodumi
Yorodumi- EMDB-20084: The Central Role of the Tail in Switching Off Myosin II in Cells -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20084 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | The Central Role of the Tail in Switching Off Myosin II in Cells | ||||||||||||
 Map data Map data | 3D reconstruction of smooth muscle myosin II molecule in the inhibited state | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Smooth Muscle / MyosinII / 10S / single particle analysis / CONTRACTILE PROTEIN | ||||||||||||
| Biological species |  | ||||||||||||
| Method | single particle reconstruction / negative staining / Resolution: 25.0 Å | ||||||||||||
 Authors Authors | Craig R / Yang SX / Lee KH / Woodhead JL / Sato O / Ikebe M | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: J Gen Physiol / Year: 2019 Journal: J Gen Physiol / Year: 2019Title: The central role of the tail in switching off 10S myosin II activity. Authors: Shixin Yang / Kyoung Hwan Lee / John L Woodhead / Osamu Sato / Mitsuo Ikebe / Roger Craig /  Abstract: Myosin II is a motor protein with two heads and an extended tail that plays an essential role in cell motility. Its active form is a polymer (myosin filament) that pulls on actin to generate motion. ...Myosin II is a motor protein with two heads and an extended tail that plays an essential role in cell motility. Its active form is a polymer (myosin filament) that pulls on actin to generate motion. Its inactive form is a monomer with a compact structure (10S sedimentation coefficient), in which the tail is folded and the two heads interact with each other, inhibiting activity. This conformation is thought to function in cells as an energy-conserving form of the molecule suitable for storage as well as transport to sites of filament assembly. The mechanism of inhibition of the compact molecule is not fully understood. We have performed a 3-D reconstruction of negatively stained 10S myosin from smooth muscle in the inhibited state using single-particle analysis. The reconstruction reveals multiple interactions between the tail and the two heads that appear to trap ATP hydrolysis products, block actin binding, hinder head phosphorylation, and prevent filament formation. Blocking these essential features of myosin function could explain the high degree of inhibition of the folded form of myosin thought to underlie its energy-conserving function in cells. The reconstruction also suggests a mechanism for unfolding when myosin is activated by phosphorylation. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20084.map.gz emd_20084.map.gz | 9.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20084-v30.xml emd-20084-v30.xml emd-20084.xml emd-20084.xml | 10.9 KB 10.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_20084.png emd_20084.png | 58.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20084 http://ftp.pdbj.org/pub/emdb/structures/EMD-20084 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20084 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20084 | HTTPS FTP |
-Validation report
| Summary document |  emd_20084_validation.pdf.gz emd_20084_validation.pdf.gz | 436.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_20084_full_validation.pdf.gz emd_20084_full_validation.pdf.gz | 436.4 KB | Display | |
| Data in XML |  emd_20084_validation.xml.gz emd_20084_validation.xml.gz | 5.4 KB | Display | |
| Data in CIF |  emd_20084_validation.cif.gz emd_20084_validation.cif.gz | 6.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20084 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20084 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20084 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20084 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_20084.map.gz / Format: CCP4 / Size: 12.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20084.map.gz / Format: CCP4 / Size: 12.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 3D reconstruction of smooth muscle myosin II molecule in the inhibited state | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.7 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Myosin II
| Entire | Name: Myosin II |
|---|---|
| Components |
|
-Supramolecule #1: Myosin II
| Supramolecule | Name: Myosin II / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Myosin II
| Macromolecule | Name: Myosin II / type: other / ID: 1 / Classification: other |
|---|---|
| Source (natural) | Organism:  |
| Sequence | String: MSQKPLSDDE KFLFVDKNFV NNPLAQADWS AKKLVWVPSE KHGFEAASIK EEKGDEVTVE LQENGKKVT LSKDDIQKMN PPKFSKVEDM AELTCLNEAS VLHNLRERYF SGLIYTYSGL F CVVINPYK QLPIYSEKII DMYKGKKRHE MPPHIYAIAD TAYRSMLQDR ...String: MSQKPLSDDE KFLFVDKNFV NNPLAQADWS AKKLVWVPSE KHGFEAASIK EEKGDEVTVE LQENGKKVT LSKDDIQKMN PPKFSKVEDM AELTCLNEAS VLHNLRERYF SGLIYTYSGL F CVVINPYK QLPIYSEKII DMYKGKKRHE MPPHIYAIAD TAYRSMLQDR EDQSILCTGE SG AGKTENT KKVIQYLAVV ASSHKGKKDT SITQGPSFSY GELEKQLLQA NPILEAFGNA KTV KNDNSS RFGKFIRINF DVTGYIVGAN IETYLLEKSR AIRQAKDERT FHIFYYLIAG ASEQ MRNDL LLEGFNNYTF LSNGHVPIPA QQDDEMFQET LEAMTIMGFT EEEQTSILRV VSSVL QLGN IVFKKERNTD QASMPDNTAA QKVCHLMGIN VTDFTRSILT PRIKVGRDVV QKAQTK EQA DFAIEALAKA KFERLFRWIL TRVNKALDKT KRQGASFLGI LDIAGFEIFE INSFEQL CI NYTNEKLQQL FNHTMFILEQ EEYQREGIEW NFIDFGLDLQ PCIELIERPT NPPGVLAL L DEECWFPKAT DTSFVEKLIQ EQGNHAKFQK SKQLKDKTEF CILHYAGKVT YNASAWLTK NMDPLNDNVT SLLNQSSDKF VADLWKDVDR IVGLDQMAKM TESSLPSASK TKKGMFRTVG QLYKEQLTK LMTTLRNTNP NFVRCIIPNH EKRAGKLDAH LVLEQLRCNG VLEGIRICRQ G FPNRIVFQ EFRQRYEILA ANAIPKGFMD GKQACILMIK ALELDPNLYR IGQSKIFFRT GV LAHLEEE RDLKITDVII AFQAQCRGYL ARKAFAKRQQ QLTAMKVIQR NCAAYLKLRN WQW WRLFTK VKPLLQVTRQ EEEMQAKDEE LQRTKERQQK AEAELKELEQ KHTQLCEEKN LLQE KLQAE TELYAEAEEM RVRLAAKKQE LEEILHEMEA RIEEEEERSQ QLQAEKKKMQ QQMLD LEEQ LEEEEAARQK LQLEKVTADG KIKKMEDDIL IMEDQNNKLT KERKLLEERV SDLTTN LAE EEEKAKNLTK LKNKHESMIS ELEVRLKKEE KSRQELEKIK RKLEGESSDL HEQIAEL QA QIAELKAQLA KKEEELQAAL ARLEDETSQK NNALKKIREL ESHISDLQED LESEKAAR N KAEKQKRDLS EELEALKTEL EDTLDTTATQ QELRAKREQE VTVLKRALEE ETRTHEAQV QEMRQKHTQA VEELTEQLEQ FKRAKANLDK TKQTLEKDNA DLANEIRSLS QAKQDVEHKK KKLEVQLQD LQSKYSDGER VRTELNEKVH KLQIEVENVT SLLNEAESKN IKLTKDVATL G SQLQDTQE LLQEETRQKL NVTTKLRQLE DDKNSLQEQL DEEVEAKQNL ERHISTLTIQ LS DSKKKLQ EFTATVETME EGKKKLQREI ESLTQQFEEK AASYDKLEKT KNRLQQELDD LVV DLDNQR QLVSNLEKKQ KKFDQMLAEE KNISSKYADE RDRAEAEARE KETKALSLAR ALEE ALEAK EELERTNKML KAEMEDLVSS KDDVGKNVHE LEKSKRTLEQ QVEEMKTQLE ELEDE LQAA EDAKLRLEVN MQAMKSQFER DLQARDEQNE EKRRQLLKQL HEHETELEDE RKQRAL AAA AKKKLEVDVK DLESQVDSAN KAREEAIKQL RKLQAQMKDY QRDLDDARAA REEIFAT AR ENEKKAKNLE AELIQLQEDL AAAERARKQA DLEKEEMAEE LASANSGRTS LQDEKRRL E ARIAQLEEEL DEEHSNIETM SDRMRKAVQQ AEQLNNELAT ERATAQKNEN ARQQLERQN KELRSKLQEM EGAVKSKFKS TIAALEAKIA SLEEQLEQEA REKQAAAKTL RQKDKKLKDA LLQVEDERK QAEQYKDQAE KGNLRLKQLK RQLEEAEEES QRINANRRKL QRELDEATES N DALGREVA ALKSKLRRGN EPVSFAPPRR SGGRRVIENA TDGGEEEIDG RDGDFNGKAS E |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Staining | Type: NEGATIVE / Material: uranyl acetate |
| Grid | Details: unspecified |
- Electron microscopy
Electron microscopy
| Microscope | FEI/PHILIPS CM120T |
|---|---|
| Image recording | Film or detector model: TVIPS TEMCAM-F224 (2k x 2k) / Average electron dose: 20.0 e/Å2 |
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
- Image processing
Image processing
| Startup model | Type of model: RANDOM CONICAL TILT |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 25.0 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 15833 |
| Initial angle assignment | Type: OTHER |
| Final angle assignment | Type: PROJECTION MATCHING |
 Movie
Movie Controller
Controller


 UCSF Chimera
UCSF Chimera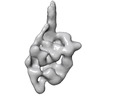





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















