[English] 日本語
 Yorodumi
Yorodumi- EMDB-19937: 6-channel super-structure of the Mechanosensitive Ion Channel of ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | 6-channel super-structure of the Mechanosensitive Ion Channel of Small Conductance 2 from Nematocida displodere | |||||||||||||||
 Map data Map data | Six homo-heptameric channels, formed by Nematocida displodere Mechanosensitive ion channel of small conductance 2, form a higher oligomeric structure resembling a six-way cross joint. | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | Membrane protein / channel protein | |||||||||||||||
| Biological species |  Nematocida displodere (fungus) Nematocida displodere (fungus) | |||||||||||||||
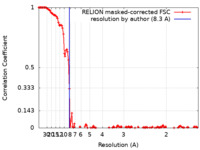
| Method | single particle reconstruction / cryo EM / Resolution: 8.3 Å | |||||||||||||||
 Authors Authors | Berg A / Barandun J / Berntsson RP-A | |||||||||||||||
| Funding support |  Sweden, European Union, 4 items Sweden, European Union, 4 items
| |||||||||||||||
 Citation Citation |  Journal: PLoS One / Year: 2024 Journal: PLoS One / Year: 2024Title: Nematocida displodere mechanosensitive ion channel of small conductance 2 assembles into a unique 6-channel super-structure in vitro. Authors: Alexandra Berg / Ronnie P-A Berntsson / Jonas Barandun /  Abstract: Mechanosensitive ion channels play an essential role in reacting to environmental signals and sustaining cell integrity by facilitating ion flux across membranes. For obligate intracellular pathogens ...Mechanosensitive ion channels play an essential role in reacting to environmental signals and sustaining cell integrity by facilitating ion flux across membranes. For obligate intracellular pathogens like microsporidia, adapting to changes in the host environment is crucial for survival and propagation. Despite representing a eukaryote of extreme genome reduction, microsporidia have expanded the gene family of mechanosensitive ion channels of small conductance (mscS) through repeated gene duplication and horizontal gene transfer. All microsporidian genomes characterized to date contain mscS genes of both eukaryotic and bacterial origin. Here, we investigated the cryo-electron microscopy structure of the bacterially derived mechanosensitive ion channel of small conductance 2 (MscS2) from Nematocida displodere, an intracellular pathogen of Caenorhabditis elegans. MscS2 is the most compact MscS-like channel known and assembles into a unique superstructure in vitro with six heptameric MscS2 channels. Individual MscS2 channels are oriented in a heterogeneous manner to one another, resembling an asymmetric, flexible six-way cross joint. Finally, we show that microsporidian MscS2 still forms a heptameric membrane channel, however the extreme compaction suggests a potential new function of this MscS-like protein. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_19937.map.gz emd_19937.map.gz | 196.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-19937-v30.xml emd-19937-v30.xml emd-19937.xml emd-19937.xml | 18.1 KB 18.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_19937_fsc.xml emd_19937_fsc.xml | 13.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_19937.png emd_19937.png | 130.6 KB | ||
| Masks |  emd_19937_msk_1.map emd_19937_msk_1.map | 216 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-19937.cif.gz emd-19937.cif.gz | 4.5 KB | ||
| Others |  emd_19937_additional_1.map.gz emd_19937_additional_1.map.gz emd_19937_half_map_1.map.gz emd_19937_half_map_1.map.gz emd_19937_half_map_2.map.gz emd_19937_half_map_2.map.gz | 199.3 MB 170.4 MB 170.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-19937 http://ftp.pdbj.org/pub/emdb/structures/EMD-19937 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19937 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19937 | HTTPS FTP |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_19937.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_19937.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Six homo-heptameric channels, formed by Nematocida displodere Mechanosensitive ion channel of small conductance 2, form a higher oligomeric structure resembling a six-way cross joint. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.8 Å | ||||||||||||||||||||||||||||||||||||
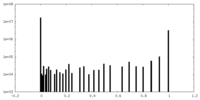
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_19937_msk_1.map emd_19937_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
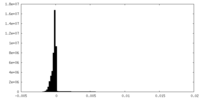
| Density Histograms |
-Additional map: #1
| File | emd_19937_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
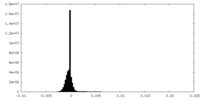
| Density Histograms |
-Half map: #2
| File | emd_19937_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
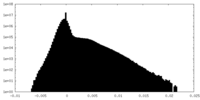
| Density Histograms |
-Half map: #1
| File | emd_19937_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Six homo-heptameric mechanosensitive ion channel of small conduct...
| Entire | Name: Six homo-heptameric mechanosensitive ion channel of small conductance 2 |
|---|---|
| Components |
|
-Supramolecule #1: Six homo-heptameric mechanosensitive ion channel of small conduct...
| Supramolecule | Name: Six homo-heptameric mechanosensitive ion channel of small conductance 2 type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Nematocida displodere (fungus) Nematocida displodere (fungus) |
| Molecular weight | Theoretical: 64 KDa |
-Supramolecule #2: Homo-heptameric mechanosensitive ion channel of small conductance 2
| Supramolecule | Name: Homo-heptameric mechanosensitive ion channel of small conductance 2 type: organelle_or_cellular_component / ID: 2 / Parent: 1 |
|---|---|
| Source (natural) | Organism:  Nematocida displodere (fungus) Nematocida displodere (fungus) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.288 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| |||||||||||||||
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Support film - Film thickness: 2 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Details: 15 mA | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Number grids imaged: 1 / Number real images: 2000 / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 4.5 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 190000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
 Movie
Movie Controller
Controller



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)