+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of Integrator subcomplex INTS5/8/15 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | RNA polymerase II transcription termination / Integrator complex assembly / transcription factors / TRANSCRIPTION | |||||||||
| Function / homology |  Function and homology information Function and homology informationsnRNA 3'-end processing / snRNA processing / INTAC complex / eye development / regulation of transcription elongation by RNA polymerase II / integrator complex / RNA polymerase II transcription initiation surveillance / RNA polymerase II transcribes snRNA genes / protein localization to chromatin / mRNA splicing, via spliceosome ...snRNA 3'-end processing / snRNA processing / INTAC complex / eye development / regulation of transcription elongation by RNA polymerase II / integrator complex / RNA polymerase II transcription initiation surveillance / RNA polymerase II transcribes snRNA genes / protein localization to chromatin / mRNA splicing, via spliceosome / brain development / chromosome / nuclear membrane / chromatin / nucleoplasm / membrane / nucleus / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Razew M / Galej WP | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2024 Journal: Mol Cell / Year: 2024Title: Structural basis of the Integrator complex assembly and association with transcription factors. Authors: Michal Razew / Angelique Fraudeau / Moritz M Pfleiderer / Romain Linares / Wojciech P Galej /  Abstract: Integrator is a multi-subunit protein complex responsible for premature transcription termination of coding and non-coding RNAs. This is achieved via two enzymatic activities, RNA endonuclease and ...Integrator is a multi-subunit protein complex responsible for premature transcription termination of coding and non-coding RNAs. This is achieved via two enzymatic activities, RNA endonuclease and protein phosphatase, acting on the promoter-proximally paused RNA polymerase Ⅱ (RNAPⅡ). Yet, it remains unclear how Integrator assembly and recruitment are regulated and what the functions of many of its core subunits are. Here, we report the structures of two human Integrator sub-complexes: INTS10/13/14/15 and INTS5/8/10/15, and an integrative model of the fully assembled Integrator bound to the RNAPⅡ paused elongating complex (PEC). An in silico protein-protein interaction screen of over 1,500 human transcription factors (TFs) identified ZNF655 as a direct interacting partner of INTS13 within the fully assembled Integrator. We propose a model wherein INTS13 acts as a platform for the recruitment of TFs that could modulate the stability of the Integrator's association at specific loci and regulate transcription attenuation of the target genes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_19872.map.gz emd_19872.map.gz | 8.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-19872-v30.xml emd-19872-v30.xml emd-19872.xml emd-19872.xml | 19.7 KB 19.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_19872.png emd_19872.png | 51.6 KB | ||
| Filedesc metadata |  emd-19872.cif.gz emd-19872.cif.gz | 7.3 KB | ||
| Others |  emd_19872_half_map_1.map.gz emd_19872_half_map_1.map.gz emd_19872_half_map_2.map.gz emd_19872_half_map_2.map.gz | 764.2 MB 764.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-19872 http://ftp.pdbj.org/pub/emdb/structures/EMD-19872 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19872 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19872 | HTTPS FTP |
-Related structure data
| Related structure data |  9ep4MC  9eocC  9eofC  9ep1C  9fa4C  9fa7C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_19872.map.gz / Format: CCP4 / Size: 824 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_19872.map.gz / Format: CCP4 / Size: 824 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.82 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_19872_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
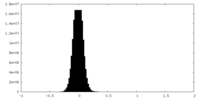
| Density Histograms |
-Half map: #1
| File | emd_19872_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
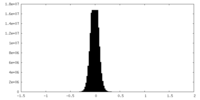
| Density Histograms |
- Sample components
Sample components
-Entire : Integrator INTS5/8/15 subcomplex
| Entire | Name: Integrator INTS5/8/15 subcomplex |
|---|---|
| Components |
|
-Supramolecule #1: Integrator INTS5/8/15 subcomplex
| Supramolecule | Name: Integrator INTS5/8/15 subcomplex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Integrator complex subunit 8
| Macromolecule | Name: Integrator complex subunit 8 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 113.219859 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MSAEAADREA ATSSRPCTPP QTCWFEFLLE ESLLEKHLRK PCPDPAPVQL IVQFLEQASK PSVNEQNQVQ PPPDNKRNRI LKLLALKVA AHLKWDLDIL EKSLSVPVLN MLLNELLCIS KVPPGTKHVD MDLATLPPTT AMAVLLYNRW AIRTIVQSSF P VKQAKPGP ...String: MSAEAADREA ATSSRPCTPP QTCWFEFLLE ESLLEKHLRK PCPDPAPVQL IVQFLEQASK PSVNEQNQVQ PPPDNKRNRI LKLLALKVA AHLKWDLDIL EKSLSVPVLN MLLNELLCIS KVPPGTKHVD MDLATLPPTT AMAVLLYNRW AIRTIVQSSF P VKQAKPGP PQLSVMNQMQ QEKELTENIL KVLKEQAADS ILVLEAALKL NKDLYVHTMR TLDLLAMEPG MVNGETESST AG LKVKTEE MQCQVCYDLG AAYFQQGSTN SAVYENAREK FFRTKELIAE IGSLSLHCTI DEKRLAGYCQ ACDVLVPSSD STS QQLTPY SQVHICLRSG NYQEVIQIFI EDNLTLSLPV QFRQSVLREL FKKAQQGNEA LDEICFKVCA CNTVRDILEG RTIS VQFNQ LFLRPNKEKI DFLLEVCSRS VNLEKASESL KGNMAAFLKN VCLGLEDLQY VFMISSHELF ITLLKDEERK LLVDQ MRKR SPRVNLCIKP VTSFYDIPAS ASVNIGQLEH QLILSVDPWR IRQILIELHG MTSERQFWTV SNKWEVPSVY SGVILG IKD NLTRDLVYIL MAKGLHCSTV KDFSHAKQLF AACLELVTEF SPKLRQVMLN EMLLLDIHTH EAGTGQAGER PPSDLIS RV RGYLEMRLPD IPLRQVIAEE CVAFMLNWRE NEYLTLQVPA FLLQSNPYVK LGQLLAATCK ELPGPKESRR TAKDLWEV V VQICSVSSQH KRGNDGRVSL IKQRESTLGI MYRSELLSFI KKLREPLVLT IILSLFVKLH NVREDIVNDI TAEHISIWP SSIPNLQSVD FEAVAITVKE LVRYTLSINP NNHSWLIIQA DIYFATNQYS AALHYYLQAG AVCSDFFNKA VPPDVYTDQV IKRMIKCCS LLNCHTQVAI LCQFLREIDY KTAFKSLQEQ NSHDAMDSYY DYIWDVTILE YLTYLHHKRG ETDKRQIAIK A IGQTELNA SNPEEVLQLA AQRRKKKFLQ AMAKLYF UniProtKB: Integrator complex subunit 8 |
-Macromolecule #2: Integrator complex subunit 5
| Macromolecule | Name: Integrator complex subunit 5 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 108.115227 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MSALCDPPGA PGPPGPAPAT HGPAPLSAQE LSQEIKAFLT GVDPILGHQL SAREHARCGL LLLRSLPPAR AAVLDHLRGV FDESVRAHL AALDETPVAG PPHLRPPPPS HVPAGGPGLE DVVQEVQQVL SEFIRANPKA WAPVISAWSI DLMGQLSSTY S GQHQRVPH ...String: MSALCDPPGA PGPPGPAPAT HGPAPLSAQE LSQEIKAFLT GVDPILGHQL SAREHARCGL LLLRSLPPAR AAVLDHLRGV FDESVRAHL AALDETPVAG PPHLRPPPPS HVPAGGPGLE DVVQEVQQVL SEFIRANPKA WAPVISAWSI DLMGQLSSTY S GQHQRVPH ATGALNELLQ LWMGCRATRT LMDIYVQCLS ALIGSCPDAC VDALLDTSVQ HSPHFDWVVA HIGSSFPGTI IS RVLSCGL KDFCVHGGAG GGAGSSGGSS SQTPSTDPFP GSPAIPAEKR VPKIASVVGI LGHLASRHGD SIRRELLRMF HDS LAGGSG GRSGDPSLQA TVPFLLQLAV MSPALLGTVS GELVDCLKPP AVLSQLQQHL QGFPREELDN MLNLAVHLVS QASG AGAYR LLQFLVDTAM PASVITTQGL AVPDTVREAC DRLIQLLLLH LQKLVHHRGG SPGEGVLGPP PPPRLVPFLD ALKNH VGEL CGETLRLERK RFLWQHQLLG LLSVYTRPSC GPEALGHLLS RARSPEELSL ATQLYAGLVV SLSGLLPLAF RSCLAR VHA GTLQPPFTAR FLRNLALLVG WEQQGGEGPA ALGAHFGESA SAHLSDLAPL LLHPEEEVAE AAASLLAICP FPSEALS PS QLLGLVRAGV HRFFASLRLH GPPGVASACQ LLTRLSQTSP AGLKAVLQLL VEGALHRGNT ELFGGQVDGD NETLSVVS A SLASASLLDT NRRHTAAVPG PGGIWSVFHA GVIGRGLKPP KFVQSRNQQE VIYNTQSLLS LLVHCCSAPG GTECGECWG APILSPEAAK AVAVTLVESV CPDAAGAELA WPPEEHARAT VERDLRIGRR FREQPLLFEL LKLVAAAPPA LCYCSVLLRG LLAALLGHW EASRHPDTTH SPWHLEASCT LVAVMAEGSL LPPALGNMHE VFSQLAPFEV RLLLLSVWGF LREHGPLPQK F IFQSERGR FIRDFSREGG GEGGPHLAVL HSVLHRNIDR LGLFSGRFQA PSPSTLLRQG T UniProtKB: Integrator complex subunit 5 |
-Macromolecule #3: Integrator complex subunit 15
| Macromolecule | Name: Integrator complex subunit 15 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 50.102402 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MSDIRHSLLR RDALSAAKEV LYHLDIYFSS QLQSAPLPIV DKGPVELLEE FVFQVPKERS AQPKRLNSLQ ELQLLEIMCN YFQEQTKDS VRQIIFSSLF SPQGNKADDS RMSLLGKLVS MAVAVCRIPV LECAASWLQR TPVVYCVRLA KALVDDYCCL V PGSIQTLK ...String: MSDIRHSLLR RDALSAAKEV LYHLDIYFSS QLQSAPLPIV DKGPVELLEE FVFQVPKERS AQPKRLNSLQ ELQLLEIMCN YFQEQTKDS VRQIIFSSLF SPQGNKADDS RMSLLGKLVS MAVAVCRIPV LECAASWLQR TPVVYCVRLA KALVDDYCCL V PGSIQTLK QIFSASPRFC CQFITSVTAL YDLSSDDLIP PMDLLEMIVT WIFEDPRLIL ITFLNTPIAA NLPIGFLELT PL VGLIRWC VKAPLAYKRK KKPPLSNGHV SNKVTKDPGV GMDRDSHLLY SKLHLSVLQV LMTLQLHLTE KNLYGRLGLI LFD HMVPLV EEINRLADEL NPLNASQEIE LSLDRLAQAL QVAMASGALL CTRDDLRTLC SRLPHNNLLQ LVISGPVQQS PHAA LPPGF YPHIHTPPLG YGAVPAHPAA HPALPTHPGH TFISGVTFPF RPIR UniProtKB: Integrator complex subunit 15 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source: LAB6 |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)