[English] 日本語
 Yorodumi
Yorodumi- EMDB-19845: Outward-open structure of human dopamine transporter bound to cocaine -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Outward-open structure of human dopamine transporter bound to cocaine | |||||||||||||||||||||
 Map data Map data | MAP SHARP | |||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||
 Keywords Keywords | neurotransmitter/sodium symporters / monoamine transporter / dopamine transporter / SLC6 / MEMBRANE PROTEIN | |||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationDefective SLC6A3 causes Parkinsonism-dystonia infantile (PKDYS) / Defective SLC6A3 causes Parkinsonism-dystonia infantile (PKDYS) / Dopamine clearance from the synaptic cleft / amine binding / dopamine uptake / adenohypophysis development / hyaloid vascular plexus regression / dopamine binding / norepinephrine:sodium symporter activity / dopamine:sodium symporter activity ...Defective SLC6A3 causes Parkinsonism-dystonia infantile (PKDYS) / Defective SLC6A3 causes Parkinsonism-dystonia infantile (PKDYS) / Dopamine clearance from the synaptic cleft / amine binding / dopamine uptake / adenohypophysis development / hyaloid vascular plexus regression / dopamine binding / norepinephrine:sodium symporter activity / dopamine:sodium symporter activity / neurotransmitter transmembrane transporter activity / regulation of dopamine metabolic process / dopamine transport / flotillin complex / dopamine catabolic process / dopaminergic synapse / monoamine transmembrane transporter activity / monoamine transport / positive regulation of multicellular organism growth / heterocyclic compound binding / SLC-mediated transport of neurotransmitters / response to iron ion / dopamine biosynthetic process / neurotransmitter transport / dopamine uptake involved in synaptic transmission / amino acid transport / response to cAMP / prepulse inhibition / lactation / axon terminus / protein phosphatase 2A binding / sodium ion transmembrane transport / response to nicotine / response to cocaine / locomotory behavior / cognition / sensory perception of smell / presynaptic membrane / protease binding / response to ethanol / postsynaptic membrane / neuron projection / membrane raft / response to xenobiotic stimulus / signaling receptor binding / axon / neuronal cell body / protein-containing complex binding / cell surface / metal ion binding / membrane / plasma membrane / cytoplasm Similarity search - Function | |||||||||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||||||||
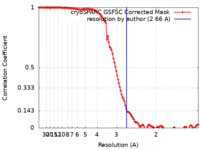
| Method | single particle reconstruction / cryo EM / Resolution: 2.66 Å | |||||||||||||||||||||
 Authors Authors | Nielsen JC / Salomon K / Kalenderoglou IE / Bargmeyer S / Pape T / Shahsavar A / Loland CJ | |||||||||||||||||||||
| Funding support |  Denmark, European Union, 6 items Denmark, European Union, 6 items
| |||||||||||||||||||||
 Citation Citation |  Journal: Nature / Year: 2024 Journal: Nature / Year: 2024Title: Structure of the human dopamine transporter in complex with cocaine. Authors: Jeppe C Nielsen / Kristine Salomon / Iris E Kalenderoglou / Sarah Bargmeyer / Tillmann Pape / Azadeh Shahsavar / Claus J Loland /  Abstract: The dopamine transporter (DAT) is crucial for regulating dopamine signalling and is the prime mediator for the rewarding and addictive effects of cocaine. As part of the neurotransmitter sodium ...The dopamine transporter (DAT) is crucial for regulating dopamine signalling and is the prime mediator for the rewarding and addictive effects of cocaine. As part of the neurotransmitter sodium symporter family, DAT uses the Na gradient across cell membranes to transport dopamine against its chemical gradient. The transport mechanism involves both intra- and extracellular gates that control substrate access to a central site. However, the molecular intricacies of this process and the inhibitory mechanism of cocaine have remained unclear. Here, we present the molecular structure of human DAT in complex with cocaine at a resolution of 2.66 Å. Our findings reveal that DAT adopts the expected LeuT-fold, posing in an outward-open conformation with cocaine bound at the central (S1) site. Notably, while an Na occupies the second Na site (Na2), the Na1 site seems to be vacant, with the side chain of Asn82 occupying the presumed Na space. This structural insight elucidates the mechanism for the cocaine inhibition of human DAT and deepens our understanding of neurotransmitter transport. By shedding light on the molecular underpinnings of how cocaine acts, our study lays a foundation for the development of targeted medications to combat addiction. | |||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_19845.map.gz emd_19845.map.gz | 157 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-19845-v30.xml emd-19845-v30.xml emd-19845.xml emd-19845.xml | 21.7 KB 21.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_19845_fsc.xml emd_19845_fsc.xml | 11.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_19845.png emd_19845.png | 81.8 KB | ||
| Masks |  emd_19845_msk_1.map emd_19845_msk_1.map | 166.4 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-19845.cif.gz emd-19845.cif.gz | 6.4 KB | ||
| Others |  emd_19845_additional_1.map.gz emd_19845_additional_1.map.gz emd_19845_half_map_1.map.gz emd_19845_half_map_1.map.gz emd_19845_half_map_2.map.gz emd_19845_half_map_2.map.gz | 82.5 MB 154.4 MB 154.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-19845 http://ftp.pdbj.org/pub/emdb/structures/EMD-19845 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19845 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19845 | HTTPS FTP |
-Related structure data
| Related structure data |  9eo4MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_19845.map.gz / Format: CCP4 / Size: 166.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_19845.map.gz / Format: CCP4 / Size: 166.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | MAP SHARP | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.725 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_19845_msk_1.map emd_19845_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: EM MAP
| File | emd_19845_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | EM MAP | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: HALF MAP B
| File | emd_19845_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | HALF MAP B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: HALF MAP A
| File | emd_19845_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | HALF MAP A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Human dopamine transporter bound to cocaine
| Entire | Name: Human dopamine transporter bound to cocaine |
|---|---|
| Components |
|
-Supramolecule #1: Human dopamine transporter bound to cocaine
| Supramolecule | Name: Human dopamine transporter bound to cocaine / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 720 KDa |
-Macromolecule #1: Sodium-dependent dopamine transporter
| Macromolecule | Name: Sodium-dependent dopamine transporter / type: protein_or_peptide / ID: 1 / Details: triple tandem GGGS linker and Twin-Strep tag / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 72.209281 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MSKSKCSVGL MSSVVAPAKE PNAVGPKEVE LILVKEQNGV QLTSSTLTNP RQSPVEAQDR ETWGKKIDFL LSVIGFAVDL ANVWRFPYL CYKNGGGAFL VPYLLFMVIA GMPLFYMELA LGQFNREGAA GVWKICPILK GVGFTVILIS LYVGFFYNVI I AWALHYLF ...String: MSKSKCSVGL MSSVVAPAKE PNAVGPKEVE LILVKEQNGV QLTSSTLTNP RQSPVEAQDR ETWGKKIDFL LSVIGFAVDL ANVWRFPYL CYKNGGGAFL VPYLLFMVIA GMPLFYMELA LGQFNREGAA GVWKICPILK GVGFTVILIS LYVGFFYNVI I AWALHYLF SSFTTELPWI HCNNSWNSPN CSDAHPGDSS GDSSGLNDTF GTTPAAEYFE RGVLHLHQSH GIDDLGPPRW QL TACLVLV IVLLYFSLWK GVKTSGKVVW ITATMPYVVL TALLLRGVTL PGAIDGIRAY LSVDFYRLCE ASVWIDAATQ VCF SLGVGF GVLIAFSSYN KFTNNCYRDA IVTTSINSLT SFSSGFVVFS FLGYMAQKHS VPIGDVAKDG PGLIFIIYPE AIAT LPLSS AWAVVFFIML LTLGIDSAMG GMESVITGLI DEFQLLHRHR ELFTLFIVLA TFLLSLFCVT NGGIYVFTLL DHFAA GTSI LFGVLIEAIG VAWFYGVGQF SDDIQQMTGQ RPSLYWRLCW KLVSPCFLLF VVVVSIVTFR PPHYGAYIFP DWANAL GWV IATSSMAMVP IYAAYKFCSL PGSFREKLAY AIAPEKDREL VDRGEVRQFT LRHWLKVGGG SGGGSGGSAW SHPQFEK GG GSGGGSGGSA WSHPQFEK UniProtKB: Sodium-dependent dopamine transporter |
-Macromolecule #2: CHOLESTEROL
| Macromolecule | Name: CHOLESTEROL / type: ligand / ID: 2 / Number of copies: 3 / Formula: CLR |
|---|---|
| Molecular weight | Theoretical: 386.654 Da |
| Chemical component information |  ChemComp-CLR: |
-Macromolecule #3: CHOLESTEROL HEMISUCCINATE
| Macromolecule | Name: CHOLESTEROL HEMISUCCINATE / type: ligand / ID: 3 / Number of copies: 1 / Formula: Y01 |
|---|---|
| Molecular weight | Theoretical: 486.726 Da |
| Chemical component information |  ChemComp-Y01: |
-Macromolecule #4: SODIUM ION
| Macromolecule | Name: SODIUM ION / type: ligand / ID: 4 / Number of copies: 1 |
|---|---|
| Molecular weight | Theoretical: 22.99 Da |
-Macromolecule #5: CHLORIDE ION
| Macromolecule | Name: CHLORIDE ION / type: ligand / ID: 5 / Number of copies: 1 / Formula: CL |
|---|---|
| Molecular weight | Theoretical: 35.453 Da |
-Macromolecule #6: COCAINE
| Macromolecule | Name: COCAINE / type: ligand / ID: 6 / Number of copies: 1 / Formula: COC |
|---|---|
| Molecular weight | Theoretical: 303.353 Da |
| Chemical component information |  ChemComp-COC: |
-Macromolecule #7: water
| Macromolecule | Name: water / type: ligand / ID: 7 / Number of copies: 4 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Pressure: 0.0001 kPa |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: TFS Selectris / Energy filter - Slit width: 5 eV |
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Number grids imaged: 1 / Number real images: 25808 / Average exposure time: 3.27 sec. / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated magnification: 170700 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.6 µm / Nominal magnification: 165000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)