+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | GroEL with bound GroTAC peptide | |||||||||
 Map data Map data | GroEL with bound GroTAC peptide EM map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | GroEL / GroTAC / PROTAC / degrader / complex / CHAPERONE | |||||||||
| Function / homology |  Function and homology information Function and homology informationGroEL-GroES complex / chaperonin ATPase / virion assembly / : / isomerase activity / ATP-dependent protein folding chaperone / response to radiation / unfolded protein binding / protein folding / response to heat ...GroEL-GroES complex / chaperonin ATPase / virion assembly / : / isomerase activity / ATP-dependent protein folding chaperone / response to radiation / unfolded protein binding / protein folding / response to heat / protein refolding / magnesium ion binding / ATP hydrolysis activity / ATP binding / identical protein binding / membrane / cytosol Similarity search - Function | |||||||||
| Biological species |  | |||||||||
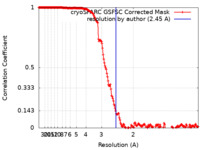
| Method | single particle reconstruction / cryo EM / Resolution: 2.45 Å | |||||||||
 Authors Authors | Wroblewski K / Izert-Nowakowska MA / Goral TK / Klimecka MM / Kmiecik S / Gorna MW | |||||||||
| Funding support |  Poland, 2 items Poland, 2 items
| |||||||||
 Citation Citation |  Journal: EMBO Rep / Year: 2025 Journal: EMBO Rep / Year: 2025Title: Targeted protein degradation in Escherichia coli using CLIPPERs. Authors: Matylda Anna Izert-Nowakowska / Maria Magdalena Klimecka / Anna Antosiewicz / Karol Wróblewski / Jakub Józef Kowalski / Katarzyna Justyna Bandyra / Tomasz Góral / Sebastian Kmiecik / ...Authors: Matylda Anna Izert-Nowakowska / Maria Magdalena Klimecka / Anna Antosiewicz / Karol Wróblewski / Jakub Józef Kowalski / Katarzyna Justyna Bandyra / Tomasz Góral / Sebastian Kmiecik / Remigiusz Adam Serwa / Maria Wiktoria Górna /  Abstract: New, universal tools for targeted protein degradation in bacteria can help to accelerate protein function studies and antimicrobial research. We describe a new method for degrading bacterial proteins ...New, universal tools for targeted protein degradation in bacteria can help to accelerate protein function studies and antimicrobial research. We describe a new method for degrading bacterial proteins using plasmid-encoded degrader peptides which deliver target proteins for degradation by a highly conserved ClpXP protease. We demonstrate the mode of action of the degraders on a challenging essential target, GroEL. The studies in bacteria are complemented by in vitro binding and structural studies. Expression of degrader peptides results in a temperature-dependent growth inhibition and depletion of GroEL levels over time. The reduction of GroEL levels is accompanied by dramatic proteome alterations. The presented method offers a new alternative approach for regulating protein levels in bacteria without genomic modifications or tag fusions. Our studies demonstrate that ClpXP is an attractive protease for the future use in bacterial-targeted protein degradation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_19687.map.gz emd_19687.map.gz | 566.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-19687-v30.xml emd-19687-v30.xml emd-19687.xml emd-19687.xml | 29.5 KB 29.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_19687_fsc.xml emd_19687_fsc.xml | 17.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_19687.png emd_19687.png | 139.7 KB | ||
| Masks |  emd_19687_msk_1.map emd_19687_msk_1.map | 600.7 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-19687.cif.gz emd-19687.cif.gz | 7.8 KB | ||
| Others |  emd_19687_additional_1.map.gz emd_19687_additional_1.map.gz emd_19687_half_map_1.map.gz emd_19687_half_map_1.map.gz emd_19687_half_map_2.map.gz emd_19687_half_map_2.map.gz | 290.6 MB 556.6 MB 556.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-19687 http://ftp.pdbj.org/pub/emdb/structures/EMD-19687 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19687 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19687 | HTTPS FTP |
-Validation report
| Summary document |  emd_19687_validation.pdf.gz emd_19687_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_19687_full_validation.pdf.gz emd_19687_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_19687_validation.xml.gz emd_19687_validation.xml.gz | 27.6 KB | Display | |
| Data in CIF |  emd_19687_validation.cif.gz emd_19687_validation.cif.gz | 36.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19687 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19687 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19687 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19687 | HTTPS FTP |
-Related structure data
| Related structure data |  8s32MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_19687.map.gz / Format: CCP4 / Size: 600.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_19687.map.gz / Format: CCP4 / Size: 600.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | GroEL with bound GroTAC peptide EM map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.5878 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_19687_msk_1.map emd_19687_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
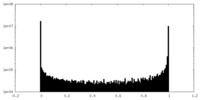
| Density Histograms |
-Additional map: Unsharpened map
| File | emd_19687_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
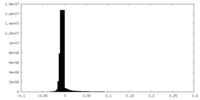
| Density Histograms |
-Half map: GroEL with bound GroTAC peptide EM half map B
| File | emd_19687_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | GroEL with bound GroTAC peptide EM half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
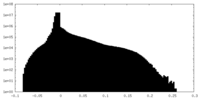
| Density Histograms |
-Half map: GroEL with bound GroTAC peptide EM half map A
| File | emd_19687_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | GroEL with bound GroTAC peptide EM half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
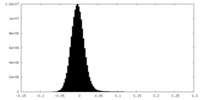
| Density Histograms |
- Sample components
Sample components
-Entire : A complex of GroEL with GroTAC peptide
| Entire | Name: A complex of GroEL with GroTAC peptide |
|---|---|
| Components |
|
-Supramolecule #1: A complex of GroEL with GroTAC peptide
| Supramolecule | Name: A complex of GroEL with GroTAC peptide / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 4 kDa/nm |
-Supramolecule #2: GroEL
| Supramolecule | Name: GroEL / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #3: GroTAC
| Supramolecule | Name: GroTAC / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
-Macromolecule #1: Chaperonin GroEL
| Macromolecule | Name: Chaperonin GroEL / type: protein_or_peptide / ID: 1 / Number of copies: 14 / Enantiomer: LEVO / EC number: chaperonin ATPase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 57.391711 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAAKDVKFGN DARVKMLRGV NVLADAVKVT LGPKGRNVVL DKSFGAPTIT KDGVSVAREI ELEDKFENMG AQMVKEVASK ANDAAGDGT TTATVLAQAI ITEGLKAVAA GMNPMDLKRG IDKAVTAAVE ELKALSVPCS DSKAIAQVGT ISANSDETVG K LIAEAMDK ...String: MAAKDVKFGN DARVKMLRGV NVLADAVKVT LGPKGRNVVL DKSFGAPTIT KDGVSVAREI ELEDKFENMG AQMVKEVASK ANDAAGDGT TTATVLAQAI ITEGLKAVAA GMNPMDLKRG IDKAVTAAVE ELKALSVPCS DSKAIAQVGT ISANSDETVG K LIAEAMDK VGKEGVITVE DGTGLQDELD VVEGMQFDRG YLSPYFINKP ETGAVELESP FILLADKKIS NIREMLPVLE AV AKAGKPL LIIAEDVEGE ALATLVVNTM RGIVKVAAVK APGFGDRRKA MLQDIATLTG GTVISEEIGM ELEKATLEDL GQA KRVVIN KDTTTIIDGV GEEAAIQGRV AQIRQQIEEA TSDYDREKLQ ERVAKLAGGV AVIKVGAATE VEMKEKKARV EDAL HATRA AVEEGVVAGG GVALIRVASK LADLRGQNED QNVGIKVALR AMEAPLRQIV LNCGEEPSVV ANTVKGGDGN YGYNA ATEE YGNMIDMGIL DPTKVTRSAL QYAASVAGLM ITTECMVTDL PKNDAADLGA AGGMGGMGGM GGMM UniProtKB: Chaperonin GroEL |
-Macromolecule #2: GroTAC
| Macromolecule | Name: GroTAC / type: protein_or_peptide / ID: 2 / Number of copies: 14 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 3.176783 KDa |
| Sequence | String: CYRGGRPALR VVK(UNK)(UNK)(UNK)SWMT TPWGFHLP |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 4 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
Details: ATP was added to the sample immediately before disposing on a grid | |||||||||||||||
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.038 kPa | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Number grids imaged: 1 / Number real images: 4672 / Average exposure time: 16.67 sec. / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 240000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | Chain - Source name: Other / Chain - Initial model type: in silico model Details: The CA model was obtained by ModelAngelo (1.09) and then subsequently rebuilt to full-atom by cg2all (1.5). |
|---|---|
| Details | The C-alpha model was obtained by two rounds of ab initio modeling using machine learning-based ModelAngelo 1.09 software. The incorrectly assigned GroEL residues were then manually corrected. The C-alpha trace of the GroTAC peptide was initially constructed by ModelAngelo, with residue identities suggested through a comparison to the 1MNF PDB structure. The reconstruction to full-atom was performed using cg2all 1.5 software. The first stage of refinement was done in Coot (0.9.8.5). The final refinement stage involved two rounds of phenix.real_space_refine (Phenix 1.21.5207). |
| Refinement | Space: REAL / Protocol: AB INITIO MODEL / Target criteria: 0.88 |
| Output model |  PDB-8s32: |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)