[English] 日本語
 Yorodumi
Yorodumi- EMDB-19489: Tobacco mosaic virus from scanning transmission electron microsco... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Tobacco mosaic virus from scanning transmission electron microscopy at CSA=2.0 mrad | |||||||||
 Map data Map data | sharp map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | tobacco mosaic virus / VIRUS | |||||||||
| Biological species |   Tobacco mosaic virus Tobacco mosaic virus | |||||||||
| Method | helical reconstruction / cryo EM / Resolution: 5.4 Å | |||||||||
 Authors Authors | Mann D / Filopoulou A / Sachse C | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: Acta Crystallogr D Struct Biol / Year: 2024 Journal: Acta Crystallogr D Struct Biol / Year: 2024Title: VitroJet: new features and case studies. Authors: Rene J M Henderikx / Daniel Mann / Aušra Domanska / Jing Dong / Saba Shahzad / Behnam Lak / Aikaterini Filopoulou / Damian Ludig / Martin Grininger / Jeffrey Momoh / Elina Laanto / Hanna M ...Authors: Rene J M Henderikx / Daniel Mann / Aušra Domanska / Jing Dong / Saba Shahzad / Behnam Lak / Aikaterini Filopoulou / Damian Ludig / Martin Grininger / Jeffrey Momoh / Elina Laanto / Hanna M Oksanen / Kyrylo Bisikalo / Pamela A Williams / Sarah J Butcher / Peter J Peters / Bart W A M M Beulen /     Abstract: Single-particle cryo-electron microscopy has become a widely adopted method in structural biology due to many recent technological advances in microscopes, detectors and image processing. Before ...Single-particle cryo-electron microscopy has become a widely adopted method in structural biology due to many recent technological advances in microscopes, detectors and image processing. Before being able to inspect a biological sample in an electron microscope, it needs to be deposited in a thin layer on a grid and rapidly frozen. The VitroJet was designed with this aim, as well as avoiding the delicate manual handling and transfer steps that occur during the conventional grid-preparation process. Since its creation, numerous technical developments have resulted in a device that is now widely utilized in multiple laboratories worldwide. It features plasma treatment, low-volume sample deposition through pin printing, optical ice-thickness measurement and cryofixation of pre-clipped Autogrids through jet vitrification. This paper presents recent technical improvements to the VitroJet and the benefits that it brings to the cryo-EM workflow. A wide variety of applications are shown: membrane proteins, nucleosomes, fatty-acid synthase, Tobacco mosaic virus, lipid nanoparticles, tick-borne encephalitis viruses and bacteriophages. These case studies illustrate the advancement of the VitroJet into an instrument that enables accurate control and reproducibility, demonstrating its suitability for time-efficient cryo-EM structure determination. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_19489.map.gz emd_19489.map.gz | 33 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-19489-v30.xml emd-19489-v30.xml emd-19489.xml emd-19489.xml | 16.1 KB 16.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_19489_fsc.xml emd_19489_fsc.xml | 8.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_19489.png emd_19489.png | 90.6 KB | ||
| Masks |  emd_19489_msk_1.map emd_19489_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-19489.cif.gz emd-19489.cif.gz | 4.7 KB | ||
| Others |  emd_19489_half_map_1.map.gz emd_19489_half_map_1.map.gz emd_19489_half_map_2.map.gz emd_19489_half_map_2.map.gz | 59.1 MB 59.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-19489 http://ftp.pdbj.org/pub/emdb/structures/EMD-19489 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19489 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19489 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_19489.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_19489.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharp map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.25 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_19489_msk_1.map emd_19489_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
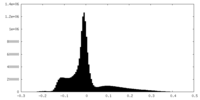
| Density Histograms |
-Half map: half map b
| File | emd_19489_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map b | ||||||||||||
| Projections & Slices |
| ||||||||||||
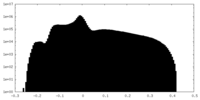
| Density Histograms |
-Half map: half map a
| File | emd_19489_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map a | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Tobacco mosaic virus
| Entire | Name:   Tobacco mosaic virus Tobacco mosaic virus |
|---|---|
| Components |
|
-Supramolecule #1: Tobacco mosaic virus
| Supramolecule | Name: Tobacco mosaic virus / type: virus / ID: 1 / Parent: 0 / NCBI-ID: 12242 / Sci species name: Tobacco mosaic virus / Virus type: VIRION / Virus isolate: OTHER / Virus enveloped: Yes / Virus empty: No |
|---|
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | helical array |
- Sample preparation
Sample preparation
| Concentration | 33 mg/mL |
|---|---|
| Buffer | pH: 7 / Details: distilled water (MilliQ) |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 278 K / Instrument: OTHER Details: VitroJet pins were cleaned with detergent and 70% EtOH in an ultrasonicator followed by 1 min drying under nitrogen stream (for 5 minutes each). TMV was pin printed at 4 degC chamber ...Details: VitroJet pins were cleaned with detergent and 70% EtOH in an ultrasonicator followed by 1 min drying under nitrogen stream (for 5 minutes each). TMV was pin printed at 4 degC chamber temperature with a 70 micrometer spiral, 15 micrometer standoff with a velocity of 5 mm per second. The value given for _em_vitrification.instrument is CRYOSOL VITROJET. This is not in a list of allowed values {'HOMEMADE PLUNGER', 'FEI VITROBOT MARK III', 'FEI VITROBOT MARK II', 'FEI VITROBOT MARK I', 'GATAN CRYOPLUNGE 3', 'LEICA PLUNGER', 'LEICA EM GP', 'REICHERT-JUNG PLUNGER', 'LEICA EM CPC', 'OTHER', 'FEI VITROBOT MARK IV', 'SPOTITON', 'EMS-002 RAPID IMMERSION FREEZER', 'LEICA KF80'} so OTHER is written into the XML file. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Details | in focus iDPC-STEM measurement using Velox software |
| Image recording | Film or detector model: OTHER / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Number real images: 103 / Average electron dose: 35.0 e/Å2 / Details: segmented TFS Panther iDPC-STEM detector |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated defocus max: 0.0 µm / Calibrated defocus min: 0.0 µm / Illumination mode: SPOT SCAN / Imaging mode: OTHER / Cs: 2.7 mm / Nominal defocus max: 0.0 µm / Nominal defocus min: 0.0 µm / Nominal magnification: 240000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)