[English] 日本語
 Yorodumi
Yorodumi- EMDB-19457: Human mitochondrial RNase Z complex with ELAC2-D550N catalytic mu... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human mitochondrial RNase Z complex with ELAC2-D550N catalytic mutant with ordered flexible arm and tRNA-Tyr precursor - (Composite model) | ||||||||||||||||||
 Map data Map data | Composite map of human mitochondrial RNase Z complex with tRNA-Tyr precursor - Class with ordered flexible arm | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | tRNA processing / RNase Z / endonuclease / mitochondrial / RNA BINDING PROTEIN | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationtRNase Z / brexanolone metabolic process / isoursodeoxycholate 7-beta-dehydrogenase (NAD+) activity / ursodeoxycholate 7-beta-dehydrogenase (NAD+) activity / 3-hydroxy-2-methylbutyryl-CoA dehydrogenase / 3-hydroxy-2-methylbutyryl-CoA dehydrogenase activity / mitochondrial tRNA methylation / tRNA (adenine9-N1)-methyltransferase / mitochondrial tRNA processing / tRNA (adenine(9)-N1)-methyltransferase activity ...tRNase Z / brexanolone metabolic process / isoursodeoxycholate 7-beta-dehydrogenase (NAD+) activity / ursodeoxycholate 7-beta-dehydrogenase (NAD+) activity / 3-hydroxy-2-methylbutyryl-CoA dehydrogenase / 3-hydroxy-2-methylbutyryl-CoA dehydrogenase activity / mitochondrial tRNA methylation / tRNA (adenine9-N1)-methyltransferase / mitochondrial tRNA processing / tRNA (adenine(9)-N1)-methyltransferase activity / mitochondrial ribonuclease P complex / mitochondrial tRNA 5'-end processing / chenodeoxycholate 7-alpha-dehydrogenase (NAD+) activity / tRNA modification in the mitochondrion / rRNA processing in the mitochondrion / tRNA processing in the mitochondrion / tRNA (guanine9-N1)-methyltransferase / tRNA (guanosine(9)-N1)-methyltransferase activity / mitochondrial tRNA 3'-end processing / 7alpha-hydroxysteroid dehydrogenase / 17-beta-hydroxysteroid dehydrogenase (NAD+) activity / cholate 7-alpha-dehydrogenase (NAD+) activity / mitochondrial RNA 5'-end processing / C21-steroid hormone metabolic process / 3'-tRNA processing endoribonuclease activity / tRNA methyltransferase complex / 3-hydroxyacyl-CoA dehydrogenase / tRNA 3'-end processing / tRNA-specific ribonuclease activity / L-isoleucine catabolic process / 3alpha(17beta)-hydroxysteroid dehydrogenase (NAD+) / tRNA-derived small RNA (tsRNA or tRNA-related fragment, tRF) biogenesis / (3S)-3-hydroxyacyl-CoA dehydrogenase (NAD+) activity / tRNA decay / : / 3alpha(or 20beta)-hydroxysteroid dehydrogenase / androstan-3-alpha,17-beta-diol dehydrogenase (NAD+) activity / testosterone dehydrogenase (NAD+) activity / positive regulation of mitochondrial translation / bile acid biosynthetic process / 17beta-estradiol 17-dehydrogenase / estradiol 17-beta-dehydrogenase [NAD(P)+] activity / Branched-chain amino acid catabolism / tRNA processing in the nucleus / estrogen metabolic process / fatty acid beta-oxidation / androgen metabolic process / mitochondrial nucleoid / RNA endonuclease activity / Mitochondrial protein degradation / Transferases; Transferring one-carbon groups; Methyltransferases / mitochondrion organization / fatty acid metabolic process / lipid metabolic process / mRNA processing / protein homotetramerization / tRNA binding / mitochondrial matrix / mitochondrion / RNA binding / nucleoplasm / metal ion binding / identical protein binding / nucleus / plasma membrane / cytosol / cytoplasm Similarity search - Function | ||||||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | ||||||||||||||||||
 Authors Authors | Bhatta A / Yu RD / Kuhle B / Hillen HS | ||||||||||||||||||
| Funding support |  Germany, European Union, 5 items Germany, European Union, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2025 Journal: Nat Struct Mol Biol / Year: 2025Title: Molecular basis of human nuclear and mitochondrial tRNA 3' processing. Authors: Arjun Bhatta / Bernhard Kuhle / Ryan D Yu / Lucas Spanaus / Katja Ditter / Katherine E Bohnsack / Hauke S Hillen /  Abstract: Eukaryotic transfer RNA (tRNA) precursors undergo sequential processing steps to become mature tRNAs. In humans, ELAC2 carries out 3' end processing of both nucleus-encoded (nu-tRNAs) and ...Eukaryotic transfer RNA (tRNA) precursors undergo sequential processing steps to become mature tRNAs. In humans, ELAC2 carries out 3' end processing of both nucleus-encoded (nu-tRNAs) and mitochondria-encoded (mt-tRNAs) tRNAs. ELAC2 is self-sufficient for processing of nu-tRNAs but requires TRMT10C and SDR5C1 to process most mt-tRNAs. Here we show that TRMT10C and SDR5C1 specifically facilitate processing of structurally degenerate mt-tRNAs lacking the canonical elbow. Structures of ELAC2 in complex with TRMT10C, SDR5C1 and two divergent mt-tRNA substrates reveal two distinct mechanisms of pre-tRNA recognition. While canonical nu-tRNAs and mt-tRNAs are recognized by direct ELAC2-RNA interactions, processing of noncanonical mt-tRNAs depends on protein-protein interactions between ELAC2 and TRMT10C. These results provide the molecular basis for tRNA 3' processing in both the nucleus and the mitochondria and explain the organelle-specific requirement for additional factors. Moreover, they suggest that TRMT10C-SDR5C1 evolved as a mitochondrial tRNA maturation platform to compensate for the structural erosion of mt-tRNAs in bilaterian animals. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_19457.map.gz emd_19457.map.gz | 90.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-19457-v30.xml emd-19457-v30.xml emd-19457.xml emd-19457.xml | 42.1 KB 42.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_19457_fsc.xml emd_19457_fsc.xml emd_19457_fsc_2.xml emd_19457_fsc_2.xml | 12 KB 12 KB | Display Display |  FSC data file FSC data file |
| Images |  emd_19457.png emd_19457.png | 47.2 KB | ||
| Masks |  emd_19457_msk_1.map emd_19457_msk_1.map | 178 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-19457.cif.gz emd-19457.cif.gz | 9.3 KB | ||
| Others |  emd_19457_additional_1.map.gz emd_19457_additional_1.map.gz emd_19457_additional_2.map.gz emd_19457_additional_2.map.gz emd_19457_additional_3.map.gz emd_19457_additional_3.map.gz emd_19457_additional_4.map.gz emd_19457_additional_4.map.gz emd_19457_additional_5.map.gz emd_19457_additional_5.map.gz emd_19457_additional_6.map.gz emd_19457_additional_6.map.gz emd_19457_half_map_1.map.gz emd_19457_half_map_1.map.gz emd_19457_half_map_2.map.gz emd_19457_half_map_2.map.gz | 88.5 MB 7 MB 89 MB 5 MB 164.9 MB 164.9 MB 165.1 MB 165 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-19457 http://ftp.pdbj.org/pub/emdb/structures/EMD-19457 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19457 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19457 | HTTPS FTP |
-Related structure data
| Related structure data |  8rr4MC  8rr1C  8rr3C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_19457.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_19457.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Composite map of human mitochondrial RNase Z complex with tRNA-Tyr precursor - Class with ordered flexible arm | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.834 Å | ||||||||||||||||||||||||||||||||||||
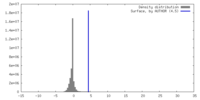
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_19457_msk_1.map emd_19457_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Focused refinement map centered on ELAC2 density
| File | emd_19457_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Focused refinement map centered on ELAC2 density | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Local resolution-filtered map for global consensus refinement
| File | emd_19457_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Local resolution-filtered map for global consensus refinement | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Consensus refinement map
| File | emd_19457_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Consensus refinement map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Local resolution filtered map for focused refinement around...
| File | emd_19457_additional_4.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Local resolution filtered map for focused refinement around ELAC2 density | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Half map 1 for focused refinement around ELAC2 density
| File | emd_19457_additional_5.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 for focused refinement around ELAC2 density | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Half map 2 for focused refinement around ELAC2 density
| File | emd_19457_additional_6.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 for focused refinement around ELAC2 density | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 2 for consensus refinement map
| File | emd_19457_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 for consensus refinement map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 1 for consensus refinement map
| File | emd_19457_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 for consensus refinement map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Human mitochondrial RNase Z complex with tRNA-Tyr precursor
| Entire | Name: Human mitochondrial RNase Z complex with tRNA-Tyr precursor |
|---|---|
| Components |
|
-Supramolecule #1: Human mitochondrial RNase Z complex with tRNA-Tyr precursor
| Supramolecule | Name: Human mitochondrial RNase Z complex with tRNA-Tyr precursor type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: 3-hydroxyacyl-CoA dehydrogenase type-2
| Macromolecule | Name: 3-hydroxyacyl-CoA dehydrogenase type-2 / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO / EC number: 3-hydroxyacyl-CoA dehydrogenase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 26.947021 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAAACRSVKG LVAVITGGAS GLGLATAERL VGQGASAVLL DLPNSGGEAQ AKKLGNNCVF APADVTSEKD VQTALALAKG KFGRVDVAV NCAGIAVASK TYNLKKGQTH TLEDFQRVLD VNLMGTFNVI RLVAGEMGQN EPDQGGQRGV IINTASVAAF E GQVGQAAY ...String: MAAACRSVKG LVAVITGGAS GLGLATAERL VGQGASAVLL DLPNSGGEAQ AKKLGNNCVF APADVTSEKD VQTALALAKG KFGRVDVAV NCAGIAVASK TYNLKKGQTH TLEDFQRVLD VNLMGTFNVI RLVAGEMGQN EPDQGGQRGV IINTASVAAF E GQVGQAAY SASKGGIVGM TLPIARDLAP IGIRVMTIAP GLFGTPLLTS LPEKVCNFLA SQVPFPSRLG DPAEYAHLVQ AI IENPFLN GEVIRLDGAI RMQP UniProtKB: 3-hydroxyacyl-CoA dehydrogenase type-2 |
-Macromolecule #2: Zinc phosphodiesterase ELAC protein 2
| Macromolecule | Name: Zinc phosphodiesterase ELAC protein 2 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO / EC number: tRNase Z |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 89.169336 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: SNAPRKDPLR HLRTREKRGP SGCSGGPNTV YLQVVAAGSR DSGAALYVFS EFNRYLFNCG EGVQRLMQEH KLKVARLDNI FLTRMHWSN VGGLSGMILT LKETGLPKCV LSGPPQLEKY LEAIKIFSGP LKGIELAVRP HSAPEYEDET MTVYQIPIHS E QRRGKHQP ...String: SNAPRKDPLR HLRTREKRGP SGCSGGPNTV YLQVVAAGSR DSGAALYVFS EFNRYLFNCG EGVQRLMQEH KLKVARLDNI FLTRMHWSN VGGLSGMILT LKETGLPKCV LSGPPQLEKY LEAIKIFSGP LKGIELAVRP HSAPEYEDET MTVYQIPIHS E QRRGKHQP WQSPERPLSR LSPERSSDSE SNENEPHLPH GVSQRRGVRD SSLVVAFICK LHLKRGNFLV LKAKEMGLPV GT AAIAPII AAVKDGKSIT HEGREILAEE LCTPPDPGAA FVVVECPDES FIQPICENAT FQRYQGKADA PVALVVHMAP ASV LVDSRY QQWMERFGPD TQHLVLNENC ASVHNLRSHK IQTQLNLIHP DIFPLLTSFR CKKEGPTLSV PMVQGECLLK YQLR PRREW QRDAIITCNP EEFIVEALQL PNFQQSVQEY RRSAQDGPAP AEKRSQYPEI IFLGTGSAIP MKIRNVSATL VNISP DTSL LLDCGEGTFG QLCRHYGDQV DRVLGTLAAV FVSHLHANHH TGLPSILLQR ERALASLGKP LHPLLVVAPN QLKAWL QQY HNQCQEVLHH ISMIPAKCLQ EGAEISSPAV ERLISSLLRT CDLEEFQTCL VRHCKHAFGC ALVHTSGWKV VYSGDTM PC EALVRMGKDA TLLIHEATLE DGLEEEAVEK THSTTSQAIS VGMRMNAEFI MLNHFSQRYA KVPLFSPNFS EKVGVAFD H MKVCFGDFPT MPKLIPPLKA LFAGDIEEME ERREKRELRQ VRAALLSREL AGGLEDGEPQ QKRAHTEEPQ AKKVRAQ UniProtKB: Zinc phosphodiesterase ELAC protein 2 |
-Macromolecule #3: tRNA methyltransferase 10 homolog C
| Macromolecule | Name: tRNA methyltransferase 10 homolog C / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO EC number: Transferases; Transferring one-carbon groups; Methyltransferases |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 37.165094 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SNAAATREFI EMWRLLGREV PEHITEEELK TLMECVSNTA KKKYLKYLYT KEKVKKARQI KKEMKAAARE EAKNIKLLET TEEDKQKNF LFLRLWDRNM DIAMGWKGAQ AMQFGQPLVF DMAYENYMKR KELQNTVSQL LESEGWNRRN VDPFHIYFCN L KIDGALHR ...String: SNAAATREFI EMWRLLGREV PEHITEEELK TLMECVSNTA KKKYLKYLYT KEKVKKARQI KKEMKAAARE EAKNIKLLET TEEDKQKNF LFLRLWDRNM DIAMGWKGAQ AMQFGQPLVF DMAYENYMKR KELQNTVSQL LESEGWNRRN VDPFHIYFCN L KIDGALHR ELVKRYQEKW DKLLLTSTEK SHVDLFPKDS IIYLTADSPN VMTTFRHDKV YVIGSFVDKS MQPGTSLAKA KR LNLATEC LPLDKYLQWE IGNKNLTLDQ MIRILLCLKN NGNWQEALQF VPKRKHTGFL EISQHSQEFI NRLKKAKT UniProtKB: tRNA methyltransferase 10 homolog C |
-Macromolecule #4: Human mitochondria tRNA-Tyr precursor with 3' trailer
| Macromolecule | Name: Human mitochondria tRNA-Tyr precursor with 3' trailer / type: rna / ID: 4 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 29.023184 KDa |
| Sequence | String: GGUAAAAUGG CUGAGUGAAG CAUUGGACUG UAAAUCUAAA GACAGGGGUU AGGCCUCUUU UUACCAGCUC CGAGGUGAUU UUCAAGCUC G GENBANK: GENBANK: MK617242.1 |
-Macromolecule #5: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 5 / Number of copies: 2 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Macromolecule #6: S-ADENOSYL-L-HOMOCYSTEINE
| Macromolecule | Name: S-ADENOSYL-L-HOMOCYSTEINE / type: ligand / ID: 6 / Number of copies: 1 / Formula: SAH |
|---|---|
| Molecular weight | Theoretical: 384.411 Da |
| Chemical component information |  ChemComp-SAH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 Component:
| |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Pretreatment - Type: GLOW DISCHARGE | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 105000 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




















 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)