+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | GPCR - G-protein complex | |||||||||
 Map data Map data | Cryo-EM map of TAS2R14 in complex with G-proteins | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Single particle cryo-EM / GPCR / G-protein / STRUCTURAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationbitter taste receptor activity / sensory perception of bitter taste / taste receptor activity / sensory perception of umami taste / detection of light stimulus involved in visual perception / sensory perception of sweet taste / detection of chemical stimulus involved in sensory perception of bitter taste / Class C/3 (Metabotropic glutamate/pheromone receptors) / phototransduction, visible light / axoneme ...bitter taste receptor activity / sensory perception of bitter taste / taste receptor activity / sensory perception of umami taste / detection of light stimulus involved in visual perception / sensory perception of sweet taste / detection of chemical stimulus involved in sensory perception of bitter taste / Class C/3 (Metabotropic glutamate/pheromone receptors) / phototransduction, visible light / axoneme / Adenylate cyclase inhibitory pathway / acrosomal vesicle / response to nicotine / G protein-coupled receptor binding / G protein-coupled receptor activity / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / G-protein beta/gamma-subunit complex binding / Olfactory Signaling Pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G-protein activation / G beta:gamma signalling through CDC42 / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / ADP signalling through P2Y purinoceptor 12 / photoreceptor disc membrane / Glucagon-type ligand receptors / Sensory perception of sweet, bitter, and umami (glutamate) taste / Adrenaline,noradrenaline inhibits insulin secretion / Vasopressin regulates renal water homeostasis via Aquaporins / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / G alpha (z) signalling events / ADP signalling through P2Y purinoceptor 1 / ADORA2B mediated anti-inflammatory cytokines production / cellular response to catecholamine stimulus / G beta:gamma signalling through PI3Kgamma / adenylate cyclase-activating dopamine receptor signaling pathway / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / GPER1 signaling / G-protein beta-subunit binding / cellular response to prostaglandin E stimulus / heterotrimeric G-protein complex / G alpha (12/13) signalling events / Inactivation, recovery and regulation of the phototransduction cascade / extracellular vesicle / sensory perception of taste / Thrombin signalling through proteinase activated receptors (PARs) / signaling receptor complex adaptor activity / retina development in camera-type eye / GTPase binding / Ca2+ pathway / fibroblast proliferation / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / G alpha (i) signalling events / G alpha (s) signalling events / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (q) signalling events / Ras protein signal transduction / Extra-nuclear estrogen signaling / cell population proliferation / apical plasma membrane / G protein-coupled receptor signaling pathway / lysosomal membrane / GTPase activity / synapse / GTP binding / protein-containing complex binding / signal transduction / protein-containing complex / extracellular exosome / metal ion binding / membrane / plasma membrane / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.03 Å | |||||||||
 Authors Authors | Matzov D / Peri L / Niv M / Shalev Benami M | |||||||||
| Funding support | European Union, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: A bitter anti-inflammatory drug binds at two distinct sites of a human bitter taste GPCR. Authors: Lior Peri / Donna Matzov / Dominic R Huxley / Alon Rainish / Fabrizio Fierro / Liel Sapir / Tara Pfeiffer / Lukas Waterloo / Harald Hübner / Yoav Peleg / Peter Gmeiner / Peter J McCormick / ...Authors: Lior Peri / Donna Matzov / Dominic R Huxley / Alon Rainish / Fabrizio Fierro / Liel Sapir / Tara Pfeiffer / Lukas Waterloo / Harald Hübner / Yoav Peleg / Peter Gmeiner / Peter J McCormick / Dorothee Weikert / Masha Y Niv / Moran Shalev-Benami /    Abstract: Bitter taste receptors (TAS2Rs), a subfamily of G-protein coupled receptors (GPCRs) expressed orally and extraorally, elicit signaling in response to a large set of tastants. Among 25 functional ...Bitter taste receptors (TAS2Rs), a subfamily of G-protein coupled receptors (GPCRs) expressed orally and extraorally, elicit signaling in response to a large set of tastants. Among 25 functional TAS2Rs encoded in the human genome, TAS2R14 is the most promiscuous, and responds to hundreds of chemically diverse ligands. Here we present the cryo-electron microscopy (cryo-EM) structure of the human TAS2R14 in complex with its signaling partner gustducin, and bound to flufenamic acid (FFA), a clinically approved nonsteroidal anti-inflammatory drug. The structure reveals an unusual binding mode, where two copies of FFA are bound at distinct pockets: one at the canonical receptor site within the trans-membrane bundle, and the other in the intracellular facet, bridging the receptor with gustducin. Together with a pocket-specific BRET-based ligand binding assay, these results illuminate bitter taste signaling and provide tools for a site-targeted compound design. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_19445.map.gz emd_19445.map.gz | 167.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-19445-v30.xml emd-19445-v30.xml emd-19445.xml emd-19445.xml | 20.7 KB 20.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_19445.png emd_19445.png | 111.5 KB | ||
| Filedesc metadata |  emd-19445.cif.gz emd-19445.cif.gz | 7.5 KB | ||
| Others |  emd_19445_additional_1.map.gz emd_19445_additional_1.map.gz | 85.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-19445 http://ftp.pdbj.org/pub/emdb/structures/EMD-19445 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19445 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19445 | HTTPS FTP |
-Related structure data
| Related structure data |  8rqlMC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_19445.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_19445.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM map of TAS2R14 in complex with G-proteins | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.826 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: cryo-EM map of TAS2R14 in complex with G-proteins - unsharpened
| File | emd_19445_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | cryo-EM map of TAS2R14 in complex with G-proteins - unsharpened | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Human Taste receptor type 2 member 14 in complex with G-proteins
| Entire | Name: Human Taste receptor type 2 member 14 in complex with G-proteins |
|---|---|
| Components |
|
-Supramolecule #1: Human Taste receptor type 2 member 14 in complex with G-proteins
| Supramolecule | Name: Human Taste receptor type 2 member 14 in complex with G-proteins type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Molecular weight | Theoretical: 118.3 kDa/nm |
-Supramolecule #2: Human Taste receptor type 2 member 14 in complex with G-proteins
| Supramolecule | Name: Human Taste receptor type 2 member 14 in complex with G-proteins type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #3: scFv16 (antibody)
| Supramolecule | Name: scFv16 (antibody) / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #5 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Taste receptor type 2 member 14
| Macromolecule | Name: Taste receptor type 2 member 14 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 36.20007 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGGVIKSIFT FVLIVEFIIG NLGNSFIALV NCIDWVKGRK ISSVDRILTA LAISRISLVW LIFGSWCVSV FFPALFATEK MFRMLTNIW TVINHFSVWL ATGLGTFYFL KIANFSNSIF LYLKWRVKKV VLVLLLVTSV FLFLNIALIN IHINASINGY R RNKTCSSD ...String: MGGVIKSIFT FVLIVEFIIG NLGNSFIALV NCIDWVKGRK ISSVDRILTA LAISRISLVW LIFGSWCVSV FFPALFATEK MFRMLTNIW TVINHFSVWL ATGLGTFYFL KIANFSNSIF LYLKWRVKKV VLVLLLVTSV FLFLNIALIN IHINASINGY R RNKTCSSD SSNFTRFSSL IVLTSTVFIF IPFTLSLAMF LLLIFSMWKH RKKMQHTVKI SGDASTKAHR GVKSVITFFL LY AIFSLSF FISVWTSERL EENLIILSQV MGMAYPSCHS CVLILGNKKL RQASLSVLLW LRYMFKDGEP SGHKEFRESS UniProtKB: Taste receptor type 2 member 14 |
-Macromolecule #2: Guanine nucleotide-binding protein G(t) subunit alpha-3
| Macromolecule | Name: Guanine nucleotide-binding protein G(t) subunit alpha-3 type: protein_or_peptide / ID: 2 Details: Mini G alpha gustducin protein,Mini G alpha gustducin protein Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 25.983604 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGSTVSAEDK AAAERSKELE KKLQEDAERD ARTVKLLLLG ADNSGKSTIV KQMKIIHGGS GGSGGTTGII ETQFSFKDLH FRMFDVGGQ RSERKKWIHC FEDVTCIIFC ADLSDYNRMH ESLHLFNSIC NHKYFSTTSI VLFLNKKDIF QEKVTKVHLS I CFPEYTGP ...String: MGSTVSAEDK AAAERSKELE KKLQEDAERD ARTVKLLLLG ADNSGKSTIV KQMKIIHGGS GGSGGTTGII ETQFSFKDLH FRMFDVGGQ RSERKKWIHC FEDVTCIIFC ADLSDYNRMH ESLHLFNSIC NHKYFSTTSI VLFLNKKDIF QEKVTKVHLS I CFPEYTGP NTFEDAGNYI KNQFLDLNLK KEDKEIYSHM TCATDTQNAK FIFDAVTDII IKENLKDCGL F UniProtKB: Guanine nucleotide-binding protein G(t) subunit alpha-3, Guanine nucleotide-binding protein G(t) subunit alpha-3 |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 39.418086 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MHHHHHHLEV LFQGPGSSGS ELDQLRQEAE QLKNQIRDAR KACADATLSQ ITNNIDPVGR IQMRTRRTLR GHLAKIYAMH WGTDSRLLV SASQDGKLII WDSYTTNKVH AIPLRSSWVM TCAYAPSGNY VACGGLDNIC SIYNLKTREG NVRVSRELAG H TGYLSCCR ...String: MHHHHHHLEV LFQGPGSSGS ELDQLRQEAE QLKNQIRDAR KACADATLSQ ITNNIDPVGR IQMRTRRTLR GHLAKIYAMH WGTDSRLLV SASQDGKLII WDSYTTNKVH AIPLRSSWVM TCAYAPSGNY VACGGLDNIC SIYNLKTREG NVRVSRELAG H TGYLSCCR FLDDNQIVTS SGDTTCALWD IETGQQTTTF TGHTGDVMSL SLAPDTRLFV SGACDASAKL WDVREGMCRQ TF TGHESDI NAICFFPNGN AFATGSDDAT CRLFDLRADQ ELMTYSHDNI ICGITSVSFS KSGRLLLAGY DDFNCNVWDA LKA DRAGVL AGHDNRVSCL GVTDDGMAVA TGSWDSFLKI WN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #4: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.861143 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFCAI L UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #5: scFv16
| Macromolecule | Name: scFv16 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 27.340482 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: DVQLVESGGG LVQPGGSRKL SCSASGFAFS SFGMHWVRQA PEKGLEWVAY ISSGSGTIYY ADTVKGRFTI SRDDPKNTLF LQMTSLRSE DTAMYYCVRS IYYYGSSPFD FWGQGTTLTV SSGGGGSGGG GSGGGGSDIV MTQATSSVPV TPGESVSISC R SSKSLLHS ...String: DVQLVESGGG LVQPGGSRKL SCSASGFAFS SFGMHWVRQA PEKGLEWVAY ISSGSGTIYY ADTVKGRFTI SRDDPKNTLF LQMTSLRSE DTAMYYCVRS IYYYGSSPFD FWGQGTTLTV SSGGGGSGGG GSGGGGSDIV MTQATSSVPV TPGESVSISC R SSKSLLHS NGNTYLYWFL QRPGQSPQLL IYRMSNLASG VPDRFSGSGS GTAFTLTISR LEAEDVGVYY CMQHLEYPLT FG AGTKLEL KGSLEVLFQ |
-Macromolecule #6: 2-[[3-(TRIFLUOROMETHYL)PHENYL]AMINO] BENZOIC ACID
| Macromolecule | Name: 2-[[3-(TRIFLUOROMETHYL)PHENYL]AMINO] BENZOIC ACID / type: ligand / ID: 6 / Number of copies: 2 / Formula: FLF |
|---|---|
| Molecular weight | Theoretical: 281.23 Da |
| Chemical component information | 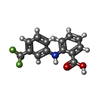 ChemComp-FLF: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 90 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 295 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 38.8 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 20.0 µm / Nominal defocus min: 8.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: INSILICO MODEL |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.03 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 127503 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller



























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)




























