[English] 日本語
 Yorodumi
Yorodumi- EMDB-1904: Labeling and Localization of the Herpes Simplex Virus Capsid Prot... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1904 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Labeling and Localization of the Herpes Simplex Virus Capsid Protein UL25 and Its Interaction with the Two Triplexes Closest to the Penton. | |||||||||
 Map data Map data | Surface rendered view of HSV-1 C-capsid where protein UL25 has been genetically tagged with GFP at amino acid 50. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | HSV-1 / capsid / UL25 / cryo-EM | |||||||||
| Biological species |   Human herpesvirus 1 strain KOS Human herpesvirus 1 strain KOS | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 13.7 Å | |||||||||
 Authors Authors | Conway JF / Cockrell SK / Copeland AM / Newcomb WW / Brown JC / Homa FL | |||||||||
 Citation Citation |  Journal: J Mol Biol / Year: 2010 Journal: J Mol Biol / Year: 2010Title: Labeling and localization of the herpes simplex virus capsid protein UL25 and its interaction with the two triplexes closest to the penton. Authors: James F Conway / Shelley K Cockrell / Anna Maria Copeland / William W Newcomb / Jay C Brown / Fred L Homa /  Abstract: The herpes simplex virus type 1 UL25 protein is one of seven viral proteins that are required for DNA cleavage and packaging. Together with UL17, UL25 forms part of an elongated molecule referred to ...The herpes simplex virus type 1 UL25 protein is one of seven viral proteins that are required for DNA cleavage and packaging. Together with UL17, UL25 forms part of an elongated molecule referred to as the C-capsid-specific component (CCSC). Five copies of the CCSC are located at each of the capsid vertices on DNA-containing capsids. To study the conformation of UL25 as it is folded on the capsid surface, we identified the sequence recognized by a UL25-specific monoclonal antibody and localized the epitope on the capsid surface by immunogold electron microscopy. The epitope mapped to amino acids 99-111 adjacent to the region of the protein (amino acids 1-50) that is required for capsid binding. In addition, cryo-EM reconstructions of C-capsids in which the green fluorescent protein (GFP) was fused within the N-terminus of UL25 localized the point of contact between UL25 and GFP. The result confirmed the modeled location of the UL25 protein in the CCSC density as the region that is distal to the penton with the N-terminus of UL25 making contact with the triplex one removed from the penton. Immunofluorescence experiments at early times during infection demonstrated that UL25-GFP was present on capsids located within the cytoplasm and adjacent to the nucleus. These results support the view that UL25 is present on incoming capsids with the capsid-binding domain of UL25 located on the surface of the mature DNA-containing capsid. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1904.map.gz emd_1904.map.gz | 530.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1904-v30.xml emd-1904-v30.xml emd-1904.xml emd-1904.xml | 8.1 KB 8.1 KB | Display Display |  EMDB header EMDB header |
| Images |  EMD-1904.png EMD-1904.png | 185.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1904 http://ftp.pdbj.org/pub/emdb/structures/EMD-1904 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1904 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1904 | HTTPS FTP |
-Validation report
| Summary document |  emd_1904_validation.pdf.gz emd_1904_validation.pdf.gz | 241.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_1904_full_validation.pdf.gz emd_1904_full_validation.pdf.gz | 240.4 KB | Display | |
| Data in XML |  emd_1904_validation.xml.gz emd_1904_validation.xml.gz | 9.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1904 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1904 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1904 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1904 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_1904.map.gz / Format: CCP4 / Size: 985.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1904.map.gz / Format: CCP4 / Size: 985.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Surface rendered view of HSV-1 C-capsid where protein UL25 has been genetically tagged with GFP at amino acid 50. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.12 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
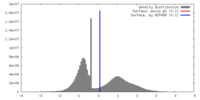
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : HSV-1 C-capsid with protein UL25 genetically labeled with GFP at ...
| Entire | Name: HSV-1 C-capsid with protein UL25 genetically labeled with GFP at amino acid 50. |
|---|---|
| Components |
|
-Supramolecule #1000: HSV-1 C-capsid with protein UL25 genetically labeled with GFP at ...
| Supramolecule | Name: HSV-1 C-capsid with protein UL25 genetically labeled with GFP at amino acid 50. type: sample / ID: 1000 / Number unique components: 1 |
|---|
-Supramolecule #1: Human herpesvirus 1 strain KOS
| Supramolecule | Name: Human herpesvirus 1 strain KOS / type: virus / ID: 1 / Name.synonym: HSV-1 / NCBI-ID: 10306 / Sci species name: Human herpesvirus 1 strain KOS / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: No / Syn species name: HSV-1 |
|---|---|
| Host (natural) | Organism:  Homo sapiens (human) / synonym: VERTEBRATES Homo sapiens (human) / synonym: VERTEBRATES |
| Virus shell | Shell ID: 1 / Diameter: 1250 Å / T number (triangulation number): 16 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 / Details: 500 mM NaCl, 10 mM Tris, 1 mM EDTA |
|---|---|
| Vitrification | Cryogen name: OTHER / Instrument: FEI VITROBOT MARK III Details: Vitrification instrument: Vitrobot mark III. Cryogen was an equal mix of ethane-propane. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: NIKON SUPER COOLSCAN 9000 / Digitization - Sampling interval: 6.35 µm / Bits/pixel: 8 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 30000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal magnification: 29000 |
| Sample stage | Specimen holder: Eucentric / Specimen holder model: GATAN LIQUID NITROGEN |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| CTF correction | Details: Phase flip each particle |
|---|---|
| Final reconstruction | Applied symmetry - Point group: I (icosahedral) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 13.7 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: AUTO3DEM |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)