[English] 日本語
 Yorodumi
Yorodumi- EMDB-19010: Plastid-encoded RNA polymerase transcription elongation complex (... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Plastid-encoded RNA polymerase transcription elongation complex (PAP2-mRNA) | |||||||||||||||
 Map data Map data | Plastid-encoded RNA polymerase transcription elongation complex (PAP2-mRNA) | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | Transcription / chloroplast / RNA polymerase / photosynthesis / GENE REGULATION | |||||||||||||||
| Biological species |  Sinapis alba (white mustard) Sinapis alba (white mustard) | |||||||||||||||
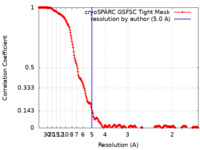
| Method | single particle reconstruction / cryo EM / Resolution: 5.0 Å | |||||||||||||||
 Authors Authors | Webster MW / Pramanick I / Vergara-Cruces A | |||||||||||||||
| Funding support |  United Kingdom, 4 items United Kingdom, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Cell / Year: 2024 Journal: Cell / Year: 2024Title: Structure of the plant plastid-encoded RNA polymerase. Authors: Ángel Vergara-Cruces / Ishika Pramanick / David Pearce / Vinod K Vogirala / Matthew J Byrne / Jason K K Low / Michael W Webster /   Abstract: Chloroplast genes encoding photosynthesis-associated proteins are predominantly transcribed by the plastid-encoded RNA polymerase (PEP). PEP is a multi-subunit complex composed of plastid-encoded ...Chloroplast genes encoding photosynthesis-associated proteins are predominantly transcribed by the plastid-encoded RNA polymerase (PEP). PEP is a multi-subunit complex composed of plastid-encoded subunits similar to bacterial RNA polymerases (RNAPs) stably bound to a set of nuclear-encoded PEP-associated proteins (PAPs). PAPs are essential to PEP activity and chloroplast biogenesis, but their roles are poorly defined. Here, we present cryoelectron microscopy (cryo-EM) structures of native 21-subunit PEP and a PEP transcription elongation complex from white mustard (Sinapis alba). We identify that PAPs encase the core polymerase, forming extensive interactions that likely promote complex assembly and stability. During elongation, PAPs interact with DNA downstream of the transcription bubble and with the nascent mRNA. The models reveal details of the superoxide dismutase, lysine methyltransferase, thioredoxin, and amino acid ligase enzymes that are subunits of PEP. Collectively, these data provide a foundation for the mechanistic understanding of chloroplast transcription and its role in plant growth and adaptation. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_19010.map.gz emd_19010.map.gz | 777 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-19010-v30.xml emd-19010-v30.xml emd-19010.xml emd-19010.xml | 12.6 KB 12.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_19010_fsc.xml emd_19010_fsc.xml | 20 KB | Display |  FSC data file FSC data file |
| Images |  emd_19010.png emd_19010.png | 79.9 KB | ||
| Filedesc metadata |  emd-19010.cif.gz emd-19010.cif.gz | 3.7 KB | ||
| Others |  emd_19010_half_map_1.map.gz emd_19010_half_map_1.map.gz emd_19010_half_map_2.map.gz emd_19010_half_map_2.map.gz | 765.4 MB 765.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-19010 http://ftp.pdbj.org/pub/emdb/structures/EMD-19010 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19010 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19010 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_19010.map.gz / Format: CCP4 / Size: 824 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_19010.map.gz / Format: CCP4 / Size: 824 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
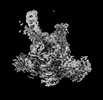
| Annotation | Plastid-encoded RNA polymerase transcription elongation complex (PAP2-mRNA) | ||||||||||||||||||||||||||||||||||||
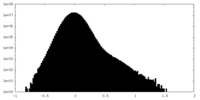
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.831 Å | ||||||||||||||||||||||||||||||||||||
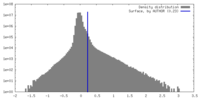
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Plastid-encoded RNA polymerase transcription elongation complex (Half map...
| File | emd_19010_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Plastid-encoded RNA polymerase transcription elongation complex (Half map A of PAP2-mRNA) | ||||||||||||
| Projections & Slices |
| ||||||||||||
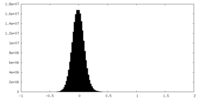
| Density Histograms |
-Half map: Plastid-encoded RNA polymerase transcription elongation complex (Half map...
| File | emd_19010_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Plastid-encoded RNA polymerase transcription elongation complex (Half map B of PAP2-mRNA) | ||||||||||||
| Projections & Slices |
| ||||||||||||
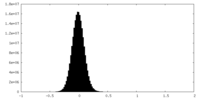
| Density Histograms |
- Sample components
Sample components
-Entire : Plastid-encoded RNA polymerase transcription elongation complex (...
| Entire | Name: Plastid-encoded RNA polymerase transcription elongation complex (PAP2-mRNA) |
|---|---|
| Components |
|
-Supramolecule #1: Plastid-encoded RNA polymerase transcription elongation complex (...
| Supramolecule | Name: Plastid-encoded RNA polymerase transcription elongation complex (PAP2-mRNA) type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Sinapis alba (white mustard) Sinapis alba (white mustard) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.6 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 40.48 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)