[English] 日本語
 Yorodumi
Yorodumi- EMDB-19000: Cryo-EM structure of coagulation factor beta-XIIa in complex with... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of coagulation factor beta-XIIa in complex with the garadacimab Fab fragment (asymmetric dimer) | |||||||||
 Map data Map data | Local resolution filtered map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Complex / coagulation / trypsin-like serine protease / hereditary angioedema (HAE) / BLOOD CLOTTING | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
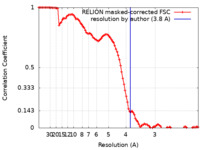
| Method | single particle reconstruction / cryo EM / Resolution: 3.8 Å | |||||||||
 Authors Authors | Drulyte I | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Structure / Year: 2024 Journal: Structure / Year: 2024Title: Structural basis for the inhibition of βFXIIa by garadacimab. Authors: Ieva Drulyte / Rajesh Ghai / Saw Yen Ow / Eugene A Kapp / Adam J Quek / Con Panousis / Michael J Wilson / Andrew D Nash / Matthias Pelzing /   Abstract: Activated FXII (FXIIa) is the principal initiator of the plasma contact system and can activate both procoagulant and proinflammatory pathways. Its activity is important in the pathophysiology of ...Activated FXII (FXIIa) is the principal initiator of the plasma contact system and can activate both procoagulant and proinflammatory pathways. Its activity is important in the pathophysiology of hereditary angioedema (HAE). Here, we describe a high-resolution cryoelectron microscopy (cryo-EM) structure of the beta-chain from FXIIa (βFXIIa) complexed with the Fab fragment of garadacimab. Garadacimab binds to βFXIIa through an unusually long CDR-H3 that inserts into the S1 pocket in a non-canonical way. This structural mechanism is likely the primary contributor to the inhibition of activated FXIIa proteolytic activity in HAE. Garadacimab Fab-βFXIIa structure also reveals critical determinants of high-affinity binding of garadacimab to activated FXIIa. Structural analysis with other bona fide FXIIa inhibitors, such as benzamidine and C1-INH, reveals a surprisingly similar mechanism of βFXIIa inhibition by garadacimab. In summary, the garadacimab Fab-βFXIIa structure provides crucial insights into its mechanism of action and delineates primary and auxiliary paratopes/epitopes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_19000.map.gz emd_19000.map.gz | 2.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-19000-v30.xml emd-19000-v30.xml emd-19000.xml emd-19000.xml | 20.3 KB 20.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_19000_fsc.xml emd_19000_fsc.xml | 10 KB | Display |  FSC data file FSC data file |
| Images |  emd_19000.png emd_19000.png | 59.4 KB | ||
| Masks |  emd_19000_msk_1.map emd_19000_msk_1.map emd_19000_msk_2.map emd_19000_msk_2.map | 83.7 MB 83.7 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-19000.cif.gz emd-19000.cif.gz | 4.8 KB | ||
| Others |  emd_19000_additional_1.map.gz emd_19000_additional_1.map.gz emd_19000_additional_2.map.gz emd_19000_additional_2.map.gz emd_19000_half_map_1.map.gz emd_19000_half_map_1.map.gz emd_19000_half_map_2.map.gz emd_19000_half_map_2.map.gz | 42.1 MB 79.1 MB 77.6 MB 77.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-19000 http://ftp.pdbj.org/pub/emdb/structures/EMD-19000 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19000 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19000 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_19000.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_19000.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Local resolution filtered map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.095 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_19000_msk_1.map emd_19000_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Mask #2
| File |  emd_19000_msk_2.map emd_19000_msk_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Unsharpened map
| File | emd_19000_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Sharpened map
| File | emd_19000_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map B
| File | emd_19000_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map A
| File | emd_19000_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Ternary complex of coagulation factor beta-XIIa with garadacimab ...
| Entire | Name: Ternary complex of coagulation factor beta-XIIa with garadacimab Fab fragment and anti-LC-lambda VHH |
|---|---|
| Components |
|
-Supramolecule #1: Ternary complex of coagulation factor beta-XIIa with garadacimab ...
| Supramolecule | Name: Ternary complex of coagulation factor beta-XIIa with garadacimab Fab fragment and anti-LC-lambda VHH type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 174 KDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL |
|---|---|
| Buffer | pH: 5.5 / Details: 10 mM Na Acetate, 100 mM NaCl pH 5.5 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
| Details | Immediately before blotting and plunge freezing, fluorinated octyl maltoside (FOM) was added to the sample to the final concentration of 0.005%-0.01% (w/v) |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: TFS Selectris / Energy filter - Slit width: 10 eV Details: Electron source E-CFEG (cold-FEG), energy filter Selectris X |
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Number grids imaged: 2 / Number real images: 4000 / Average electron dose: 50.0 e/Å2 / Details: 2000 with 0.005% FOM and 2000 with 0.01% FOM |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.25 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 165000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)