+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Human SHMT1 | |||||||||
 Map data Map data | Cryo-EM structure of SHMT1. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Riboregulation / Serine / Glycine Metabolism / 1 carbon metablism / Moonlighting protein / RNA BINDING PROTEIN / TRANSFERASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationcellular response to tetrahydrofolate / Carnitine synthesis / carnitine biosynthetic process / purine nucleobase biosynthetic process / serine binding / L-serine catabolic process / glycine metabolic process / L-serine metabolic process / aldehyde-lyase activity / glycine hydroxymethyltransferase ...cellular response to tetrahydrofolate / Carnitine synthesis / carnitine biosynthetic process / purine nucleobase biosynthetic process / serine binding / L-serine catabolic process / glycine metabolic process / L-serine metabolic process / aldehyde-lyase activity / glycine hydroxymethyltransferase / glycine hydroxymethyltransferase activity / glycine biosynthetic process from serine / Metabolism of folate and pterines / tetrahydrofolate metabolic process / tetrahydrofolate interconversion / dTMP biosynthetic process / small molecule binding / folic acid metabolic process / mRNA regulatory element binding translation repressor activity / cellular response to leukemia inhibitory factor / mRNA 5'-UTR binding / pyridoxal phosphate binding / protein homotetramerization / negative regulation of translation / protein homodimerization activity / mitochondrion / extracellular exosome / nucleoplasm / identical protein binding / nucleus / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.29 Å | |||||||||
 Authors Authors | Spizzichino S / Marabelli C / Bharadwaj A / Jakobi AJ / Chaves-Sanjuan A / Giardina G / Bolognesi M / Cutruzzola F | |||||||||
| Funding support |  Italy, 2 items Italy, 2 items
| |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2024 Journal: Mol Cell / Year: 2024Title: Structure-based mechanism of riboregulation of the metabolic enzyme SHMT1. Authors: Sharon Spizzichino / Federica Di Fonzo / Chiara Marabelli / Angela Tramonti / Antonio Chaves-Sanjuan / Alessia Parroni / Giovanna Boumis / Francesca Romana Liberati / Alessio Paone / Linda ...Authors: Sharon Spizzichino / Federica Di Fonzo / Chiara Marabelli / Angela Tramonti / Antonio Chaves-Sanjuan / Alessia Parroni / Giovanna Boumis / Francesca Romana Liberati / Alessio Paone / Linda Celeste Montemiglio / Matteo Ardini / Arjen J Jakobi / Alok Bharadwaj / Paolo Swuec / Gian Gaetano Tartaglia / Alessandro Paiardini / Roberto Contestabile / Antonello Mai / Dante Rotili / Francesco Fiorentino / Alberto Macone / Alessandra Giorgi / Giancarlo Tria / Serena Rinaldo / Martino Bolognesi / Giorgio Giardina / Francesca Cutruzzolà /   Abstract: RNA can directly control protein activity in a process called riboregulation; only a few mechanisms of riboregulation have been described in detail, none of which have been characterized on ...RNA can directly control protein activity in a process called riboregulation; only a few mechanisms of riboregulation have been described in detail, none of which have been characterized on structural grounds. Here, we present a comprehensive structural, functional, and phylogenetic analysis of riboregulation of cytosolic serine hydroxymethyltransferase (SHMT1), the enzyme interconverting serine and glycine in one-carbon metabolism. We have determined the cryoelectron microscopy (cryo-EM) structure of human SHMT1 in its free- and RNA-bound states, and we show that the RNA modulator competes with polyglutamylated folates and acts as an allosteric switch, selectively altering the enzyme's reactivity vs. serine. In addition, we identify the tetrameric assembly and a flap structural motif as key structural elements necessary for binding of RNA to eukaryotic SHMT1. The results presented here suggest that riboregulation may have played a role in evolution of eukaryotic SHMT1 and in compartmentalization of one-carbon metabolism. Our findings provide insights for RNA-based therapeutic strategies targeting this cancer-linked metabolic pathway. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18973.map.gz emd_18973.map.gz | 1.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18973-v30.xml emd-18973-v30.xml emd-18973.xml emd-18973.xml | 20.5 KB 20.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_18973.png emd_18973.png | 34.2 KB | ||
| Filedesc metadata |  emd-18973.cif.gz emd-18973.cif.gz | 6.7 KB | ||
| Others |  emd_18973_half_map_1.map.gz emd_18973_half_map_1.map.gz emd_18973_half_map_2.map.gz emd_18973_half_map_2.map.gz | 194.4 MB 194.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18973 http://ftp.pdbj.org/pub/emdb/structures/EMD-18973 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18973 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18973 | HTTPS FTP |
-Validation report
| Summary document |  emd_18973_validation.pdf.gz emd_18973_validation.pdf.gz | 673.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_18973_full_validation.pdf.gz emd_18973_full_validation.pdf.gz | 673.5 KB | Display | |
| Data in XML |  emd_18973_validation.xml.gz emd_18973_validation.xml.gz | 16 KB | Display | |
| Data in CIF |  emd_18973_validation.cif.gz emd_18973_validation.cif.gz | 18.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18973 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18973 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18973 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18973 | HTTPS FTP |
-Related structure data
| Related structure data |  8r7hMC  8a11C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_18973.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18973.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM structure of SHMT1. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.889 Å | ||||||||||||||||||||||||||||||||||||
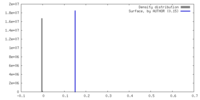
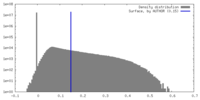
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Cryo-EM structure of SHMT1. Half map 2
| File | emd_18973_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM structure of SHMT1. Half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
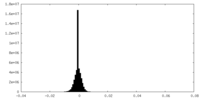
| Density Histograms |
-Half map: Cryo-EM structure of SHMT1. Half map 1
| File | emd_18973_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM structure of SHMT1. Half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
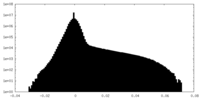
| Density Histograms |
- Sample components
Sample components
-Entire : human SHMT1
| Entire | Name: human SHMT1 |
|---|---|
| Components |
|
-Supramolecule #1: human SHMT1
| Supramolecule | Name: human SHMT1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all / Details: SHMT1= homotetramer of 53kDa subunits |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 210 KDa |
-Macromolecule #1: Serine hydroxymethyltransferase, cytosolic
| Macromolecule | Name: Serine hydroxymethyltransferase, cytosolic / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: glycine hydroxymethyltransferase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 53.662914 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GSHMTMPVNG AHKDADLWSS HDKMLAQPLK DSDVEVYNII KKESNRQRVG LELIASENFA SRAVLEALGS CLNNKYSEGY PGQRYYGGT EFIDELETLC QKRALQAYKL DPQCWGVNVQ PYSGSPANFA VYTALVEPHG RIMGLDLPDG GHLTHGFMTD K KKISATSI ...String: GSHMTMPVNG AHKDADLWSS HDKMLAQPLK DSDVEVYNII KKESNRQRVG LELIASENFA SRAVLEALGS CLNNKYSEGY PGQRYYGGT EFIDELETLC QKRALQAYKL DPQCWGVNVQ PYSGSPANFA VYTALVEPHG RIMGLDLPDG GHLTHGFMTD K KKISATSI FFESMPYKVN PDTGYINYDQ LEENARLFHP KLIIAGTSCY SRNLEYARLR KIADENGAYL MADMAHISGL VA AGVVPSP FEHCHVVTTT TH(LLP)TLRGCRA GMIFYRKGVK SVDPKTGKEI LYNLESLINS AVFPGLQGGP HNHAIAGVA VALKQAMTLE FKVYQHQVVA NCRALSEALT ELGYKIVTGG SDNHLILVDL RSKGTDGGRA EKVLEACSIA CNKNTCPGDR SALRPSGLR LGTPALTSRG LLEKDFQKVA HFIHRGIELT LQIQSDTGVR ATLKEFKERL AGDKYQAAVQ ALREEVESFA S LFPLPGLP DF UniProtKB: Serine hydroxymethyltransferase, cytosolic |
-Macromolecule #2: Serine hydroxymethyltransferase, cytosolic
| Macromolecule | Name: Serine hydroxymethyltransferase, cytosolic / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 53.434797 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GSHMTMPVNG AHKDADLWSS HDKMLAQPLK DSDVEVYNII KKESNRQRVG LELIASENFA SRAVLEALGS CLNNKYSEGY PGQRYYGGT EFIDELETLC QKRALQAYKL DPQCWGVNVQ PYSGSPANFA VYTALVEPHG RIMGLDLPDG GHLTHGFMTD K KKISATSI ...String: GSHMTMPVNG AHKDADLWSS HDKMLAQPLK DSDVEVYNII KKESNRQRVG LELIASENFA SRAVLEALGS CLNNKYSEGY PGQRYYGGT EFIDELETLC QKRALQAYKL DPQCWGVNVQ PYSGSPANFA VYTALVEPHG RIMGLDLPDG GHLTHGFMTD K KKISATSI FFESMPYKVN PDTGYINYDQ LEENARLFHP KLIIAGTSCY SRNLEYARLR KIADENGAYL MADMAHISGL VA AGVVPSP FEHCHVVTTT THKTLRGCRA GMIFYRKGVK SVDPKTGKEI LYNLESLINS AVFPGLQGGP HNHAIAGVAV ALK QAMTLE FKVYQHQVVA NCRALSEALT ELGYKIVTGG SDNHLILVDL RSKGTDGGRA EKVLEACSIA CNKNTCPGDR SALR PSGLR LGTPALTSRG LLEKDFQKVA HFIHRGIELT LQIQSDTGVR ATLKEFKERL AGDKYQAAVQ ALREEVESFA SLFPL PGLP DF UniProtKB: Serine hydroxymethyltransferase, cytosolic |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.3 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.2 Component:
| |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV / Details: blotted for 4 seconds before plunging. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 5450 / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 120000 |
| Sample stage | Specimen holder model: OTHER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)