[English] 日本語
 Yorodumi
Yorodumi- EMDB-18947: 1918 H1N1 Viral polymerase heterotrimer in complex with 4 repeat ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | 1918 H1N1 Viral polymerase heterotrimer in complex with 4 repeat serine-5 phosphorylated PolII peptide with ordered PB2 C-terminal domains | ||||||||||||||||||||||||
 Map data Map data | Unsharpened map | ||||||||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||||||||
 Keywords Keywords | influenza / polymerase / PolII-CTD / VIRAL PROTEIN | ||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationcap snatching / viral transcription / symbiont-mediated suppression of host mRNA transcription via inhibition of RNA polymerase II activity / host cell mitochondrion / 7-methylguanosine mRNA capping / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of MAVS activity / virion component / endonuclease activity / Hydrolases; Acting on ester bonds / host cell cytoplasm ...cap snatching / viral transcription / symbiont-mediated suppression of host mRNA transcription via inhibition of RNA polymerase II activity / host cell mitochondrion / 7-methylguanosine mRNA capping / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of MAVS activity / virion component / endonuclease activity / Hydrolases; Acting on ester bonds / host cell cytoplasm / symbiont-mediated suppression of host gene expression / hydrolase activity / viral translational frameshifting / RNA-directed RNA polymerase / viral RNA genome replication / nucleotide binding / RNA-directed RNA polymerase activity / DNA-templated transcription / host cell nucleus / RNA binding / metal ion binding Similarity search - Function | ||||||||||||||||||||||||
| Biological species |  Influenza A virus (A/Brevig Mission/1/1918(H1N1)) / Influenza A virus (A/Brevig Mission/1/1918(H1N1)) /  Homo sapiens (human) Homo sapiens (human) | ||||||||||||||||||||||||
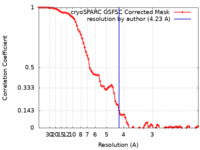
| Method | single particle reconstruction / cryo EM / Resolution: 4.23 Å | ||||||||||||||||||||||||
 Authors Authors | Keown JR / Carrique L / Fodor E / Grimes JM | ||||||||||||||||||||||||
| Funding support |  United Kingdom, 7 items United Kingdom, 7 items
| ||||||||||||||||||||||||
 Citation Citation |  Journal: J Virol / Year: 2024 Journal: J Virol / Year: 2024Title: Structural and functional characterization of the interaction between the influenza A virus RNA polymerase and the CTD of host RNA polymerase II. Authors: Jeremy Keown / Alaa Baazaoui / Marek Šebesta / Richard Štefl / Loïc Carrique / Ervin Fodor / Jonathan M Grimes /  Abstract: Influenza A viruses, causing seasonal epidemics and occasional pandemics, rely on interactions with host proteins for their RNA genome transcription and replication. The viral RNA polymerase utilizes ...Influenza A viruses, causing seasonal epidemics and occasional pandemics, rely on interactions with host proteins for their RNA genome transcription and replication. The viral RNA polymerase utilizes host RNA polymerase II (Pol II) and interacts with the serine 5 phosphorylated (pS5) C-terminal domain (CTD) of Pol II to initiate transcription. Our study, using single-particle electron cryomicroscopy (cryo-EM), reveals the structure of the 1918 pandemic influenza A virus polymerase bound to a synthetic pS5 CTD peptide composed of four heptad repeats mimicking the 52 heptad repeat mammalian Pol II CTD. The structure shows that the CTD peptide binds at the C-terminal domain of the PA viral polymerase subunit (PA-C) and reveals a previously unobserved position of the 627 domain of the PB2 subunit near the CTD. We identify crucial residues of the CTD peptide that mediate interactions with positively charged cavities on PA-C, explaining the preference of the viral polymerase for pS5 CTD. Functional analysis of mutants targeting the CTD-binding site within PA-C reveals reduced transcriptional function or defects in replication, highlighting the multifunctional role of PA-C in viral RNA synthesis. Our study provides insights into the structural and functional aspects of the influenza virus polymerase-host Pol II interaction and identifies a target for antiviral development.IMPORTANCEUnderstanding the intricate interactions between influenza A viruses and host proteins is crucial for developing targeted antiviral strategies. This study employs advanced imaging techniques to uncover the structural nuances of the 1918 pandemic influenza A virus polymerase bound to a specific host protein, shedding light on the vital process of viral RNA synthesis. The study identifies key amino acid residues in the influenza polymerase involved in binding host polymerase II (Pol II) and highlights their role in both viral transcription and genome replication. These findings not only deepen our understanding of the influenza virus life cycle but also pinpoint a potential target for antiviral development. By elucidating the structural and functional aspects of the influenza virus polymerase-host Pol II interaction, this research provides a foundation for designing interventions to disrupt viral replication and transcription, offering promising avenues for future antiviral therapies. | ||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18947.map.gz emd_18947.map.gz | 32 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18947-v30.xml emd-18947-v30.xml emd-18947.xml emd-18947.xml | 29 KB 29 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_18947_fsc.xml emd_18947_fsc.xml | 8.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_18947.png emd_18947.png | 36.1 KB | ||
| Filedesc metadata |  emd-18947.cif.gz emd-18947.cif.gz | 8.9 KB | ||
| Others |  emd_18947_additional_1.map.gz emd_18947_additional_1.map.gz emd_18947_half_map_1.map.gz emd_18947_half_map_1.map.gz emd_18947_half_map_2.map.gz emd_18947_half_map_2.map.gz | 59.4 MB 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18947 http://ftp.pdbj.org/pub/emdb/structures/EMD-18947 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18947 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18947 | HTTPS FTP |
-Related structure data
| Related structure data |  8r65MC  8r60C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_18947.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18947.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.05 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Map processed using deepEMhancer wide target with a...
| File | emd_18947_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map processed using deepEMhancer wide target with a single map as input (not halfmaps) | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half-map A
| File | emd_18947_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map_A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half-map B
| File | emd_18947_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map_B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : 1918 H1N1 Viral polymerase heterotrimer in complex with 4 repeat ...
| Entire | Name: 1918 H1N1 Viral polymerase heterotrimer in complex with 4 repeat serine-5 phosphorylated PolII peptide with ordered PB2 C-terminal domains |
|---|---|
| Components |
|
-Supramolecule #1: 1918 H1N1 Viral polymerase heterotrimer in complex with 4 repeat ...
| Supramolecule | Name: 1918 H1N1 Viral polymerase heterotrimer in complex with 4 repeat serine-5 phosphorylated PolII peptide with ordered PB2 C-terminal domains type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#6 |
|---|---|
| Source (natural) | Organism:  Influenza A virus (A/Brevig Mission/1/1918(H1N1)) Influenza A virus (A/Brevig Mission/1/1918(H1N1)) |
-Macromolecule #1: Polymerase acidic protein
| Macromolecule | Name: Polymerase acidic protein / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Influenza A virus (A/Brevig Mission/1/1918(H1N1)) Influenza A virus (A/Brevig Mission/1/1918(H1N1)) |
| Molecular weight | Theoretical: 82.663383 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MEDFVRQCFN PMIVELAEKA MKEYGEDLKI ETNKFAAICT HLEVCFMYSD FHFINERGES IIVESGDPNA LLKHRFEIIE GRDRTMAWT VVNSICNTTG AEKPKFLPAL YDYKENRFIE IGVTRREVHI YYLEKANKIK SEKTHIHIFS FTGEEMATKA D YTLDEESR ...String: MEDFVRQCFN PMIVELAEKA MKEYGEDLKI ETNKFAAICT HLEVCFMYSD FHFINERGES IIVESGDPNA LLKHRFEIIE GRDRTMAWT VVNSICNTTG AEKPKFLPAL YDYKENRFIE IGVTRREVHI YYLEKANKIK SEKTHIHIFS FTGEEMATKA D YTLDEESR ARIKTRLFTI RQEMASRGLW DSFRQSERGE ETIEERFEIT GTMRRLADQS LPPNFSSLEN FRAYVDGFEP NG YIEGKLS QMSKEVNARI EPFLKTTPRP LRLPDGPPCS QRSKFLLMDA LKLSIEDPSH EGEGIPLYDA IKCMRTFFGW KEP NVVKPH EKGINPNYLL AWKQVLAELQ DIENEEKIPK TKNMKKTSQL KWALGENMAP EKVDFDDCKD VSDLKQYDSD EPEL RSLAS WIQSEFNKAC ELTDSSWIEL DEIGEDVAPI EHIASMRRNY FTAEVSHCRA TEYIMKGVYI NTALLNASCA AMDDF QLIP MISKCRTKEG RRKTNLYGFI IKGRSHLRND TDVVNFVSME FSLTDPRLEP HKWEKYCVLE IGDMLLRSAI GQVSRP MFL YVRTNGTSKI KMKWGMEMRR CLLQSLQQIE SMIEAESSVK EKDMTKEFFE NKSETWPIGE SPKGVEEGSI GKVCRTL LA KSVFNSLYAS PQLEGFSAES RKLLLIVQAL RDNLEPGTFD LGGLYEAIEE CLINDPWVLL NASWFNSFLT HALR UniProtKB: Polymerase acidic protein |
-Macromolecule #2: Polymerase basic protein 2
| Macromolecule | Name: Polymerase basic protein 2 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Influenza A virus (A/Brevig Mission/1/1918(H1N1)) Influenza A virus (A/Brevig Mission/1/1918(H1N1)) |
| Molecular weight | Theoretical: 102.377219 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MERIKELRDL MSQSRTREIL TKTTVDHMAI IKKYTSGRQE KNPALRMKWM MAMKYPITAD KRIMEMIPER NEQGQTLWSK TNDAGSDRV MVSPLAVTWW NRNGPTTSAV HYPKIYKTYF EKVERLKHGT FGPVHFRNQV KIRRRVDINP GHADLSAKEA Q DVIMEVVF ...String: MERIKELRDL MSQSRTREIL TKTTVDHMAI IKKYTSGRQE KNPALRMKWM MAMKYPITAD KRIMEMIPER NEQGQTLWSK TNDAGSDRV MVSPLAVTWW NRNGPTTSAV HYPKIYKTYF EKVERLKHGT FGPVHFRNQV KIRRRVDINP GHADLSAKEA Q DVIMEVVF PNEVGARILT SESQLTITKE KKEELQDCKI SPLMVAYMLE RELVRKTRFL PVAGGTSSVY IEVLHLTQGT CW EQMYTPG GEVRNDDVDQ SLIIAARNIV RRATVSADPL ASLLEMCHST QIGGIRMVDI LRQNPTEEQA VDICKAAMGL RIS SSFSFG GFTFKRTSGS SVKREEEVLT GNLQTLKIRV HEGYEEFTMV GRRATAILRK ATRRLIQLIV SGRDEQSIAE AIIV AMVFS QEDCMIKAVR GDLNFVNRAN QRLNPMHQLL RHFQKDAKVL FQNWGIEPID NVMGMIGILP DMTPSTEMSM RGVRV SKMG VDEYSSTERV VVSIDRFLRV RDQRGNVLLS PEEVSETQGT EKLTITYSSS MMWEVNGPES VLVNTYQWII RNWETV KIQ WSQNPTMLYN KMEFEPFQSL VPKAARGQYS GFVRTLFQQM RDVLGTFDTV QIIKLLPFAA APPKQSRMQF SSLTVNV RG SGMRILVRGN SPVFNYNKAT KRLTVLGKDA GALTEDPDEG TAGVESAVLR GFLILGKEDR RYGPALSINE LSNLAKGE K ANVLIGQGDV VLVMKRKRDS SILTDSQTAT KRIRMAINEN LYFQGELKTA ALAQHDEAVD NKFNKEQQNA FYEILHLPN LNEEQRNAFI QSLKDDPSQS ANLLAEAKKL NDAQAPKVDN KFNKEQQNAF YEILHLPNLN EEQRNAFIQS LKADPSQSAN LLAEAKKLN GAQAPKVDAN SAGKST UniProtKB: Polymerase basic protein 2 |
-Macromolecule #5: RNA polymerase II 4 repeat peptide with serine5 phosphorylation
| Macromolecule | Name: RNA polymerase II 4 repeat peptide with serine5 phosphorylation type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 3.216893 KDa |
| Sequence | String: YSPT(SEP)PSYSP T(SEP)PSYSPT(SEP)P SYSPT(SEP)PS |
-Macromolecule #6: RNA-directed RNA polymerase catalytic subunit
| Macromolecule | Name: RNA-directed RNA polymerase catalytic subunit / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO / EC number: RNA-directed RNA polymerase |
|---|---|
| Source (natural) | Organism:  Influenza A virus (A/Brevig Mission/1/1918(H1N1)) Influenza A virus (A/Brevig Mission/1/1918(H1N1)) |
| Molecular weight | Theoretical: 86.625211 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MDVNPTLLFL KVPAQNAIST TFPYTGDPPY SHGTGTGYTM DTVNRTHQYS EKGRWTTNTE TGAPQLNPID GPLPEDNEPS GYAQTDCVL EAMAFLEESH PGIFENSCLE TMEVVQQTRV DKLTQGRQTY DWTLNRNQPA ATALANTIEV FRSNGLTANE S GRLIDFLK ...String: MDVNPTLLFL KVPAQNAIST TFPYTGDPPY SHGTGTGYTM DTVNRTHQYS EKGRWTTNTE TGAPQLNPID GPLPEDNEPS GYAQTDCVL EAMAFLEESH PGIFENSCLE TMEVVQQTRV DKLTQGRQTY DWTLNRNQPA ATALANTIEV FRSNGLTANE S GRLIDFLK DVMESMDKEE MEITTHFQRK RRVRDNMTKK MVTQRTIGKK KQRLNKRSYL IRALTLNTMT KDAERGKLKR RA IATPGMQ IRGFVYFVET LARSICEKLE QSGLPVGGNE KKAKLANVVR KMMTNSQDTE LSFTITGDNT KWNENQNPRM FLA MITYIT RNQPEWFRNV LSIAPIMFSN KMARLGKGYM FESKSMKLRT QIPAEMLASI DLKYFNDSTR KKIEKIRPLL IDGT ASLSP GMMMGMFNML STVLGVSILN LGQKRYTKTT YWWDGLQSSD DFALIVNAPN HEGIQAGVDR FYRTCKLLGI NMSKK KSYI NRTGTFEFTS FFYRYGFVAN FSMELPSFGV SGINESADMS IGVTVIKNNM INNDLGPATA QMALQLFIKD YRYTYR CHR GDTQIQTRRS FEIKKLWEQT RSKAGLLVSD GGPNLYNIRN LHIPEVCLKW ELMDEDYQGR LCNPLNPFVS HKEIESV NN AVMMPAHGPA KNMEYDAVAT THSWIPKRNR SILNTSQRGI LEDEQMYQKC CNLFEKFFPS SSYRRPVGIS SMVEAMVS R ARIDARIDFE SGRIKKEEFA EIMKICSTIE ELRRQK UniProtKB: RNA-directed RNA polymerase catalytic subunit |
-Macromolecule #3: RNA (5'-R(P*AP*GP*UP*AP*GP*AP*AP*AP*CP*AP*AP*GP*GP*CP*C)-3')
| Macromolecule | Name: RNA (5'-R(P*AP*GP*UP*AP*GP*AP*AP*AP*CP*AP*AP*GP*GP*CP*C)-3') type: rna / ID: 3 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Influenza A virus (A/Brevig Mission/1/1918(H1N1)) Influenza A virus (A/Brevig Mission/1/1918(H1N1)) |
| Molecular weight | Theoretical: 4.862017 KDa |
| Sequence | String: AGUAGAAACA AGGCC |
-Macromolecule #4: RNA (5'-R(P*GP*GP*CP*CP*UP*GP*CP*U)-3')
| Macromolecule | Name: RNA (5'-R(P*GP*GP*CP*CP*UP*GP*CP*U)-3') / type: rna / ID: 4 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Influenza A virus (A/Brevig Mission/1/1918(H1N1)) Influenza A virus (A/Brevig Mission/1/1918(H1N1)) |
| Molecular weight | Theoretical: 5.335125 KDa |
| Sequence | String: GGCCUGCUUU UGCUAUU |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.3 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.6 Component:
| |||||||||||||||
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Details | The model from 8R60 was used as a starting point. The PB2 C-terminal domains (7NHX) were split into Cap-binding domain and 627/NLS domains which were rigid body fit into the density. Seperate models were manually linked |
| Refinement | Protocol: RIGID BODY FIT |
| Output model |  PDB-8r65: |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)