[English] 日本語
 Yorodumi
Yorodumi- EMDB-18826: In situ sub-tomogram average of the E. coli 70S ribosome obtained... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | In situ sub-tomogram average of the E. coli 70S ribosome obtained using honeycomb gold supports | |||||||||
 Map data Map data | A sub-tomogram average of the 70S ribosome with E-site tRNA bound, obtained from FIB-milled vertically trapped Escherichia coli cells. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ribosome / tomography / sub-tomogram averaging | |||||||||
| Biological species |  | |||||||||
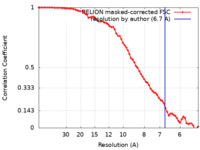
| Method | subtomogram averaging / cryo EM / Resolution: 6.7 Å | |||||||||
 Authors Authors | Hale VL / Hooker JA / Russo CJ / Lowe J | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: J Struct Biol / Year: 2024 Journal: J Struct Biol / Year: 2024Title: Honeycomb gold specimen supports enabling orthogonal focussed ion beam-milling of elongated cells for cryo-ET. Authors: Victoria L Hale / James Hooker / Christopher J Russo / Jan Löwe /  Abstract: Cryo-focussed ion beam (FIB)-milling is a powerful technique that opens up thick, cellular specimens to high-resolution structural analysis by electron cryotomography (cryo-ET). FIB-milled lamellae ...Cryo-focussed ion beam (FIB)-milling is a powerful technique that opens up thick, cellular specimens to high-resolution structural analysis by electron cryotomography (cryo-ET). FIB-milled lamellae can be produced from cells on grids, or cut from thicker, high-pressure frozen specimens. However, these approaches can put geometrical constraints on the specimen that may be unhelpful, particularly when imaging structures within the cell that have a very defined orientation. For example, plunge frozen rod-shaped bacteria orient parallel to the plane of the grid, yet the Z-ring, a filamentous structure of the tubulin-like protein FtsZ and the key organiser of bacterial division, runs around the circumference of the cell such that it is perpendicular to the imaging plane. It is therefore difficult or impractical to image many complete rings with current technologies. To circumvent this problem, we have fabricated monolithic gold specimen supports with a regular array of cylindrical wells in a honeycomb geometry, which trap bacteria in a vertical orientation. These supports, which we call "honeycomb gold discs", replace standard EM grids and when combined with FIB-milling enable the production of lamellae containing cross-sections through cells. The resulting lamellae are more stable and resistant to breakage and charging than conventional lamellae. The design of the honeycomb discs can be modified according to need and so will also enable cryo-ET and cryo-EM imaging of other specimens in otherwise difficult to obtain orientations. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18826.map.gz emd_18826.map.gz | 26 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18826-v30.xml emd-18826-v30.xml emd-18826.xml emd-18826.xml | 14.9 KB 14.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_18826_fsc.xml emd_18826_fsc.xml | 7 KB | Display |  FSC data file FSC data file |
| Images |  emd_18826.png emd_18826.png | 70 KB | ||
| Filedesc metadata |  emd-18826.cif.gz emd-18826.cif.gz | 4.6 KB | ||
| Others |  emd_18826_half_map_1.map.gz emd_18826_half_map_1.map.gz emd_18826_half_map_2.map.gz emd_18826_half_map_2.map.gz | 14.7 MB 14.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18826 http://ftp.pdbj.org/pub/emdb/structures/EMD-18826 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18826 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18826 | HTTPS FTP |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_18826.map.gz / Format: CCP4 / Size: 28.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18826.map.gz / Format: CCP4 / Size: 28.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | A sub-tomogram average of the 70S ribosome with E-site tRNA bound, obtained from FIB-milled vertically trapped Escherichia coli cells. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.659 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_18826_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
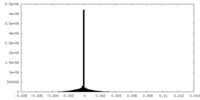
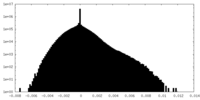
| Density Histograms |
-Half map: #1
| File | emd_18826_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
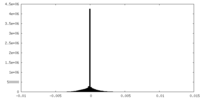
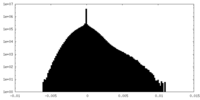
| Density Histograms |
- Sample components
Sample components
-Entire : 70S ribosome with E-site tRNA
| Entire | Name: 70S ribosome with E-site tRNA |
|---|---|
| Components |
|
-Supramolecule #1: 70S ribosome with E-site tRNA
| Supramolecule | Name: 70S ribosome with E-site tRNA / type: complex / ID: 1 / Parent: 0 / Details: Native ribosomes |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 2.5 MDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 7.7 |
|---|---|
| Grid | Material: GOLD / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 240 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.025 kPa / Details: 30 mA, 2 minutes per side |
| Vitrification | Cryogen name: ETHANE / Instrument: HOMEMADE PLUNGER / Details: Manually back-blotted at room temperature. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4088 pixel / Number real images: 5 / Average exposure time: 0.3 sec. / Average electron dose: 3.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 6.0 µm / Nominal defocus min: 2.0 µm / Nominal magnification: 33000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)