[English] 日本語
 Yorodumi
Yorodumi- EMDB-18777: Single particle cryo-EM co-structure of Klebsiella pneumoniae Acr... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Single particle cryo-EM co-structure of Klebsiella pneumoniae AcrB with the BDM91288 efflux pump inhibitor at 3.42 Angstrom resolution | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Efflux Pump Inhibitor / Transmembrane Binding / Antibiotic Resistance / TRANSPORT PROTEIN | |||||||||
| Function / homology | :  Function and homology information Function and homology information | |||||||||
| Biological species |  Klebsiella pneumoniae subsp. pneumoniae DSM 30104 = JCM 1662 = NBRC 14940 (bacteria) Klebsiella pneumoniae subsp. pneumoniae DSM 30104 = JCM 1662 = NBRC 14940 (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.42 Å | |||||||||
 Authors Authors | Boernsen C / Mueller RT / Pos KM / Frangakis AS | |||||||||
| Funding support |  Germany, Germany,  France, 2 items France, 2 items
| |||||||||
 Citation Citation |  Journal: To be published Journal: To be publishedTitle: Single particle cryo-EM co-structure of Klebsiella pneumoniae AcrB with the BDM91288 efflux pump inhibitor at 3.42 Angstrom resolution Authors: Boernsen C / Mueller RT | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18777.map.gz emd_18777.map.gz | 107.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18777-v30.xml emd-18777-v30.xml emd-18777.xml emd-18777.xml | 17.5 KB 17.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_18777_fsc.xml emd_18777_fsc.xml | 13.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_18777.png emd_18777.png | 131.3 KB | ||
| Masks |  emd_18777_msk_1.map emd_18777_msk_1.map | 209.3 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-18777.cif.gz emd-18777.cif.gz | 6.5 KB | ||
| Others |  emd_18777_half_map_1.map.gz emd_18777_half_map_1.map.gz emd_18777_half_map_2.map.gz emd_18777_half_map_2.map.gz | 194 MB 194 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18777 http://ftp.pdbj.org/pub/emdb/structures/EMD-18777 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18777 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18777 | HTTPS FTP |
-Related structure data
| Related structure data |  8qzlMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_18777.map.gz / Format: CCP4 / Size: 209.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18777.map.gz / Format: CCP4 / Size: 209.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.837 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_18777_msk_1.map emd_18777_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_18777_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_18777_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Trimeric Klebsiella pneumoniae AcrB with BDM91288 efflux pump inh...
| Entire | Name: Trimeric Klebsiella pneumoniae AcrB with BDM91288 efflux pump inhibitor |
|---|---|
| Components |
|
-Supramolecule #1: Trimeric Klebsiella pneumoniae AcrB with BDM91288 efflux pump inh...
| Supramolecule | Name: Trimeric Klebsiella pneumoniae AcrB with BDM91288 efflux pump inhibitor type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Klebsiella pneumoniae subsp. pneumoniae DSM 30104 = JCM 1662 = NBRC 14940 (bacteria) Klebsiella pneumoniae subsp. pneumoniae DSM 30104 = JCM 1662 = NBRC 14940 (bacteria)Strain: FDAARGOS_775 |
-Macromolecule #1: Efflux pump membrane transporter
| Macromolecule | Name: Efflux pump membrane transporter / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Klebsiella pneumoniae subsp. pneumoniae DSM 30104 = JCM 1662 = NBRC 14940 (bacteria) Klebsiella pneumoniae subsp. pneumoniae DSM 30104 = JCM 1662 = NBRC 14940 (bacteria)Strain: FDAARGOS_775 |
| Molecular weight | Theoretical: 114.489945 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MPNFFIDRPI FAWVIAIIIM LAGGLSILKL PVAQYPTIAP PAISITAMYP GADAETVQNT VTQVIEQNMN GIDHLMYMSS NGDSTGTAT ITLTFESGTD PDIAQVQVQN KLALATPLLP QEVQQQGISV EKASSSFLMV VGVINTNGTM NQDDISDYVA A NMKDPISR ...String: MPNFFIDRPI FAWVIAIIIM LAGGLSILKL PVAQYPTIAP PAISITAMYP GADAETVQNT VTQVIEQNMN GIDHLMYMSS NGDSTGTAT ITLTFESGTD PDIAQVQVQN KLALATPLLP QEVQQQGISV EKASSSFLMV VGVINTNGTM NQDDISDYVA A NMKDPISR TSGVGDVQLF GSQYAMRIWM DPNKLNNFQL TPVDVISALK AQNAQVAAGQ LGGTPPVKGQ QLNASIIAQT RL TNTEEFG NILLKVNQDG SQVRLRDVAK IELGGESYDV VAKFNGQPAS GLGIKLATGA NALDTANAIR AELAKMEPFF PSG MKIVYP YDTTPFVKIS IHEVVKTLVE AIILVFLVMY LFLQNFRATL IPTIAVPVVL LGTFAVLAAF GFSINTLTMF GMVL AIGLL VDDAIVVVEN VERVMAEEGL PPKEATRKSM GQIQGALVGI AMVLSAVFIP MAFFGGSTGA IYRQFSITIV SAMAL SVLV ALILTPALCA TMLKPIQKGS HGATTGFFGW FNRMFDKSTH HYTDSVGNIL RSTGRYLVLY LIIVVGMAWL FVRLPS SFL PDEDQGVFLS MAQLPAGATQ ERTQKVLDEM TNYYLTKEKD NVESVFAVNG FGFAGRGQNT GIAFVSLKDW SQRPGEE NK VEAITARAMG YFSQIKDAMV FAFNLPAIVE LGTATGFDFE LIDQGGLGHE KLTQARNQLF GMVAQHPDVL TGVRPNGL E DTPQFKIDID QEKAQALGVS ISDINTTLGA AWGGSYVNDF IDRGRVKKVY IMSEAKYRML PEDIGKWYVR GSDGQMVPF SAFSTSRWEY GSPRLERYNG LPSLEILGQA APGKSTGEAM ALMEELAGKL PSGIGYDWTG MSYQERLSGN QAPALYAISL IVVFLCLAA LYESWSIPFS VMLVVPLGVV GALLAATFRG LTNDVYFQVG LLTTIGLSAK NAILIVEFAK DLMEKEGKGL I EATLEAVR MRLRPILMTS LAFILGVMPL VISSGAGSGA QNAVGTGVMG GMVTATILAI FFVPVFFVVV RRRFSKKTED IE HSHQVEH HLEHHHHHH UniProtKB: UNIPROTKB: J2LW81 |
-Macromolecule #2: 3-chloranyl-2,6-di(piperazin-4-ium-1-yl)quinoline
| Macromolecule | Name: 3-chloranyl-2,6-di(piperazin-4-ium-1-yl)quinoline / type: ligand / ID: 2 / Number of copies: 2 / Formula: WQW |
|---|---|
| Molecular weight | Theoretical: 333.859 Da |
| Chemical component information | 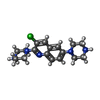 ChemComp-WQW: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 4.2 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||
| Grid | Material: COPPER | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum SE / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 8487 / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated defocus max: 3.5 µm / Calibrated defocus min: 0.8 µm / Calibrated magnification: 130000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-8qzl: |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)













































