[English] 日本語
 Yorodumi
Yorodumi- EMDB-18697: Subtomogram average of Ebola virus nucleocapsid obtained from cry... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Subtomogram average of Ebola virus nucleocapsid obtained from cryo-FIB milled Ebola virus infected Huh7 cells at 22 hours post infection | |||||||||
 Map data Map data | Subtomogram average Ebola condensed nucleocapsid from Ebola virus infected cells (low pass filtered to 21A resolution) | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Ebola virus nucleocapsid / VIRUS | |||||||||
| Biological species |  | |||||||||
| Method | subtomogram averaging / cryo EM / Resolution: 21.0 Å | |||||||||
 Authors Authors | Vallbracht M / Chlanda P | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2025 Journal: Cell / Year: 2025Title: Nucleocapsid assembly drives Ebola viral factory maturation and dispersion. Authors: Melina Vallbracht / Bianca S Bodmer / Konstantin Fischer / Jana Makroczyova / Sophie L Winter / Lisa Wendt / Moritz Wachsmuth-Melm / Thomas Hoenen / Petr Chlanda /  Abstract: Replication and genome encapsidation of many negative-sense RNA viruses take place in virus-induced membraneless organelles termed viral factories (VFs). Although liquid properties of VFs are ...Replication and genome encapsidation of many negative-sense RNA viruses take place in virus-induced membraneless organelles termed viral factories (VFs). Although liquid properties of VFs are believed to control the transition from genome replication to nucleocapsid (NC) assembly, VF maturation and interactions with the cellular environment remain elusive. Here, we apply in situ cryo-correlative light and electron tomography to follow NC assembly and changes in VF morphology and their liquid properties during Ebola virus infection. We show that viral NCs transition from loosely packed helical assemblies in early VFs to compact cylinders that arrange into highly organized parallel bundles later in infection. Early VFs associate with intermediate filaments and are devoid of other host material but become progressively accessible to cellular components. Our data suggest that this process is coupled to VF solidification, loss of sphericity, and dispersion and promotes cytoplasmic exposure of NCs to facilitate their transport to budding sites. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18697.map.gz emd_18697.map.gz | 58.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18697-v30.xml emd-18697-v30.xml emd-18697.xml emd-18697.xml | 14.8 KB 14.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_18697_fsc.xml emd_18697_fsc.xml | 11.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_18697.png emd_18697.png | 109.7 KB | ||
| Masks |  emd_18697_msk_1.map emd_18697_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-18697.cif.gz emd-18697.cif.gz | 4.6 KB | ||
| Others |  emd_18697_half_map_1.map.gz emd_18697_half_map_1.map.gz emd_18697_half_map_2.map.gz emd_18697_half_map_2.map.gz | 58.9 MB 59 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18697 http://ftp.pdbj.org/pub/emdb/structures/EMD-18697 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18697 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18697 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_18697.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18697.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Subtomogram average Ebola condensed nucleocapsid from Ebola virus infected cells (low pass filtered to 21A resolution) | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.671 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_18697_msk_1.map emd_18697_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
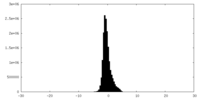
| Density Histograms |
-Half map: Subtomogram average half map of Ebola condensed nucleocapsid
| File | emd_18697_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Subtomogram average half map of Ebola condensed nucleocapsid | ||||||||||||
| Projections & Slices |
| ||||||||||||
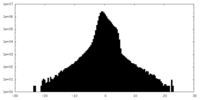
| Density Histograms |
-Half map: Subtomogram average half map of Ebola condensed nucleocapsid
| File | emd_18697_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Subtomogram average half map of Ebola condensed nucleocapsid | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Ebola virus - Mayinga, Zaire, 1976
| Entire | Name:  |
|---|---|
| Components |
|
-Supramolecule #1: Ebola virus - Mayinga, Zaire, 1976
| Supramolecule | Name: Ebola virus - Mayinga, Zaire, 1976 / type: virus / ID: 1 / Parent: 0 / NCBI-ID: 128952 / Sci species name: Ebola virus - Mayinga, Zaire, 1976 / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: Yes / Virus empty: No |
|---|---|
| Virus shell | Shell ID: 1 / Name: Nucleocapsid / Diameter: 500.0 Å |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: Quantifoil / Material: GOLD / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 3.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 4.0 µm / Nominal defocus min: 2.5 µm / Nominal magnification: 33000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)