[English] 日本語
 Yorodumi
Yorodumi- EMDB-18687: Cryo-EM Structure of Regulated CBS Domain-Containing Pyrophosphat... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM Structure of Regulated CBS Domain-Containing Pyrophosphatase without One Catalytic Domain | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | cystathionine beta-synthase domain / DHH-domain / domain swapping / HYDROLASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationinorganic diphosphatase / inorganic diphosphate phosphatase activity / metal ion binding / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Desulfitobacterium hafniense DCB-2 (bacteria) Desulfitobacterium hafniense DCB-2 (bacteria) | |||||||||
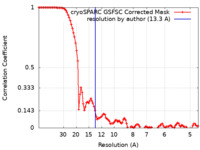
| Method | single particle reconstruction / cryo EM / Resolution: 13.3 Å | |||||||||
 Authors Authors | Moiseenko AV / Anashkin VA / Zamakhov IM / Sokolova OS / Baykov AA | |||||||||
| Funding support |  Russian Federation, 1 items Russian Federation, 1 items
| |||||||||
 Citation Citation |  Journal: Int J Mol Sci / Year: 2023 Journal: Int J Mol Sci / Year: 2023Title: The Structure and Nucleotide-Binding Characteristics of Regulated Cystathionine β-Synthase Domain-Containing Pyrophosphatase without One Catalytic Domain. Authors: Ilya M Zamakhov / Viktor A Anashkin / Andrey V Moiseenko / Victor N Orlov / Natalia N Vorobyeva / Olga S Sokolova / Alexander A Baykov /   Abstract: Regulatory adenine nucleotide-binding cystathionine β-synthase (CBS) domains are widespread in proteins; however, information on the mechanism of their modulating effects on protein function is ...Regulatory adenine nucleotide-binding cystathionine β-synthase (CBS) domains are widespread in proteins; however, information on the mechanism of their modulating effects on protein function is scarce. The difficulty in obtaining structural data for such proteins is ascribed to their unusual flexibility and propensity to form higher-order oligomeric structures. In this study, we deleted the most movable domain from the catalytic part of a CBS domain-containing bacterial inorganic pyrophosphatase (CBS-PPase) and characterized the deletion variant both structurally and functionally. The truncated CBS-PPase was inactive but retained the homotetrameric structure of the full-size enzyme and its ability to bind a fluorescent AMP analog (inhibitor) and diadenosine tetraphosphate (activator) with the same or greater affinity. The deletion stabilized the protein structure against thermal unfolding, suggesting that the deleted domain destabilizes the structure in the full-size protein. A "linear" 3D structure with an unusual type of domain swapping predicted for the truncated CBS-PPase by Alphafold2 was confirmed by single-particle electron microscopy. The results suggest a dual role for the CBS domains in CBS-PPase regulation: they allow for enzyme tetramerization, which impedes the motion of one catalytic domain, and bind adenine nucleotides to mitigate or aggravate this effect. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18687.map.gz emd_18687.map.gz | 51.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18687-v30.xml emd-18687-v30.xml emd-18687.xml emd-18687.xml | 17.4 KB 17.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_18687_fsc.xml emd_18687_fsc.xml | 10 KB | Display |  FSC data file FSC data file |
| Images |  emd_18687.png emd_18687.png | 23.1 KB | ||
| Filedesc metadata |  emd-18687.cif.gz emd-18687.cif.gz | 5.4 KB | ||
| Others |  emd_18687_half_map_1.map.gz emd_18687_half_map_1.map.gz emd_18687_half_map_2.map.gz emd_18687_half_map_2.map.gz | 95.6 MB 95.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18687 http://ftp.pdbj.org/pub/emdb/structures/EMD-18687 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18687 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18687 | HTTPS FTP |
-Validation report
| Summary document |  emd_18687_validation.pdf.gz emd_18687_validation.pdf.gz | 653.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_18687_full_validation.pdf.gz emd_18687_full_validation.pdf.gz | 653.5 KB | Display | |
| Data in XML |  emd_18687_validation.xml.gz emd_18687_validation.xml.gz | 17.9 KB | Display | |
| Data in CIF |  emd_18687_validation.cif.gz emd_18687_validation.cif.gz | 23.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18687 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18687 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18687 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18687 | HTTPS FTP |
-Related structure data
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_18687.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18687.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.33 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_18687_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_18687_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Homotetrameric CBS-PPase_delta_DHHA2-domain
| Entire | Name: Homotetrameric CBS-PPase_delta_DHHA2-domain |
|---|---|
| Components |
|
-Supramolecule #1: Homotetrameric CBS-PPase_delta_DHHA2-domain
| Supramolecule | Name: Homotetrameric CBS-PPase_delta_DHHA2-domain / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: 2,5 mg/ml 0.1 M MOPS/KOH, 2 mM MgCl2, 150 mM KCl, pH 7.2 |
|---|---|
| Source (natural) | Organism:  Desulfitobacterium hafniense DCB-2 (bacteria) Desulfitobacterium hafniense DCB-2 (bacteria) |
-Macromolecule #1: pyrophosphatase containing cystathionine beta-synthase domains an...
| Macromolecule | Name: pyrophosphatase containing cystathionine beta-synthase domains and lacking one catalytic domain type: protein_or_peptide / ID: 1 / Details: CBS-PPase without DHHA2-domain. / Enantiomer: LEVO / EC number: inorganic diphosphatase |
|---|---|
| Source (natural) | Organism:  Desulfitobacterium hafniense DCB-2 (bacteria) / Strain: DCB-2 Desulfitobacterium hafniense DCB-2 (bacteria) / Strain: DCB-2 |
| Recombinant expression | Organism:  |
| Sequence | String: MSKKIHVVGH RNPDTDSICA AIAYARLKQR LGMDHVIPYR AGKINRETEF VLNAFGVEAP ELLKDLHLR VKDMLNGPIP AVKPRTSLLE AWKIMKESNQ KTLPVVDHTE QMIGMITVGD L SGSYIESM ADHELESLHI PVENVIRTLN GRLLVGSAEQ DLKGNVYVGA ...String: MSKKIHVVGH RNPDTDSICA AIAYARLKQR LGMDHVIPYR AGKINRETEF VLNAFGVEAP ELLKDLHLR VKDMLNGPIP AVKPRTSLLE AWKIMKESNQ KTLPVVDHTE QMIGMITVGD L SGSYIESM ADHELESLHI PVENVIRTLN GRLLVGSAEQ DLKGNVYVGA MRHETLEAYV GN GNIVLMG DRQTAQESVL RQGVSALILT GGATLAPELE DLARQQNCVV ISVPYDTFTA ARL LPMTAP VQNAMKTEGI VAFNEDDLIS EVKEKMLETR FRNYPVLDEQ GKVVGLISRY HLLS LSRKQ VILVDHNEFG QAVVGTDQAQ ILEVVDHHRV GGIQTGEPIL FRNEPVGSTC TIVAK CYRD WDVVPEKAIA GIMLGAILSD TVIFKSPTCT EIDRENATYL ASIAGVDPKE FGVQMF KAA UniProtKB: inorganic diphosphatase |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.5 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.2 Component:
| ||||||||||||
| Grid | Model: EMS Lacey Carbon / Mesh: 300 / Support film - Material: CARBON / Support film - topology: LACEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.026000000000000002 kPa | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 277 K / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 2100 |
|---|---|
| Temperature | Min: 97.0 K / Max: 103.0 K |
| Image recording | Film or detector model: DIRECT ELECTRON DE-20 (5k x 3k) / Detector mode: INTEGRATING / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source: LAB6 |
| Electron optics | Calibrated defocus max: 3.3000000000000003 µm / Calibrated defocus min: 0.6 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 1.4 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 2.0 µm |
| Sample stage | Specimen holder model: JEOL / Cooling holder cryogen: NITROGEN |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)