[English] 日本語
 Yorodumi
Yorodumi- EMDB-18635: Cryo-EM structure of human SLC15A4 dimer in outward open state in LMNG -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
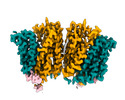
| Title | Cryo-EM structure of human SLC15A4 dimer in outward open state in LMNG | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Solute carrier / transporter / SLC / MFS / major facilitator superfamily / lysosome / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationhistidine transport / mast cell homeostasis / L-histidine transmembrane export from vacuole / L-histidine transmembrane transporter activity / peptidoglycan transmembrane transporter activity / Proton/oligopeptide cotransporters / positive regulation of toll-like receptor 8 signaling pathway / peptidoglycan transport / positive regulation of toll-like receptor 7 signaling pathway / regulation of isotype switching to IgG isotypes ...histidine transport / mast cell homeostasis / L-histidine transmembrane export from vacuole / L-histidine transmembrane transporter activity / peptidoglycan transmembrane transporter activity / Proton/oligopeptide cotransporters / positive regulation of toll-like receptor 8 signaling pathway / peptidoglycan transport / positive regulation of toll-like receptor 7 signaling pathway / regulation of isotype switching to IgG isotypes / SLC15A4:TASL-dependent IRF5 activation / positive regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway / dipeptide import across plasma membrane / positive regulation of toll-like receptor 9 signaling pathway / peptide:proton symporter activity / dipeptide transmembrane transporter activity / regulation of nucleotide-binding domain, leucine rich repeat containing receptor signaling pathway / positive regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway / positive regulation of innate immune response / endolysosome membrane / specific granule membrane / monoatomic ion transport / protein transport / early endosome membrane / innate immune response / lysosomal membrane / Neutrophil degranulation / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.81 Å | |||||||||
 Authors Authors | Rehan S / Matsuoka R / Jazayeri A / Duerr KL | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Cryo-EM structure of human SLC15A4 dimer in outward open state in LMNG Authors: Rehan S / Matsuoka R / Jazayeri A / Duerr KL | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18635.map.gz emd_18635.map.gz | 26.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18635-v30.xml emd-18635-v30.xml emd-18635.xml emd-18635.xml | 17 KB 17 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_18635.png emd_18635.png | 97.6 KB | ||
| Masks |  emd_18635_msk_1.map emd_18635_msk_1.map | 52.7 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-18635.cif.gz emd-18635.cif.gz | 5.9 KB | ||
| Others |  emd_18635_additional_1.map.gz emd_18635_additional_1.map.gz emd_18635_half_map_1.map.gz emd_18635_half_map_1.map.gz emd_18635_half_map_2.map.gz emd_18635_half_map_2.map.gz | 51.1 MB 48.7 MB 48.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18635 http://ftp.pdbj.org/pub/emdb/structures/EMD-18635 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18635 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18635 | HTTPS FTP |
-Validation report
| Summary document |  emd_18635_validation.pdf.gz emd_18635_validation.pdf.gz | 1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_18635_full_validation.pdf.gz emd_18635_full_validation.pdf.gz | 1 MB | Display | |
| Data in XML |  emd_18635_validation.xml.gz emd_18635_validation.xml.gz | 11.8 KB | Display | |
| Data in CIF |  emd_18635_validation.cif.gz emd_18635_validation.cif.gz | 13.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18635 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18635 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18635 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18635 | HTTPS FTP |
-Related structure data
| Related structure data |  8qslMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_18635.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18635.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.045 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_18635_msk_1.map emd_18635_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Sharpened map
| File | emd_18635_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_18635_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_18635_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : SLC15A4 in detergent
| Entire | Name: SLC15A4 in detergent |
|---|---|
| Components |
|
-Supramolecule #1: SLC15A4 in detergent
| Supramolecule | Name: SLC15A4 in detergent / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Solute carrier family 15 member 4
| Macromolecule | Name: Solute carrier family 15 member 4 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 62.004035 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MEGSGGGAGE RAPAAGARRA AAAAAAAGAF AGRRAACGAV LLTELLERAA FYGITSNLVL FLNGAPFCWE GAQASEALLL FMGLTYLGS PFGGWLADAR LGRARAILLS LALYLLGMLA FPLLAAPATR AALCGSARLL NCTAPGPDAA ARCCSPATFA G LVLVGLGV ...String: MEGSGGGAGE RAPAAGARRA AAAAAAAGAF AGRRAACGAV LLTELLERAA FYGITSNLVL FLNGAPFCWE GAQASEALLL FMGLTYLGS PFGGWLADAR LGRARAILLS LALYLLGMLA FPLLAAPATR AALCGSARLL NCTAPGPDAA ARCCSPATFA G LVLVGLGV ATVKANITPF GADQVKDRGP EATRRFFNWF YWSINLGAIL SLGGIAYIQQ NVSFVTGYAI PTVCVGLAFV VF LCGQSVF ITKPPDGSAF TDMFKILTYS CCSQKRSGER QSNGEGIGVF QQSSKQSLFD SCKMSHGGPF TEEKVEDVKA LVK IVPVFL ALIPYWTVYF QMQTTYVLQS LHLRIPEISN ITTTPHTLPA AWLTMFDAVL ILLLIPLKDK LVDPILRRHG LLPS SLKRI AVGMFFVMCS AFAAGILESK RLNLVKEKTI NQTIGNVVYH AADLSLWWQV PQYLLIGISE IFASIAGLEF AYSAA PKSM QSAIMGLFFF FSGVGSFVGS GLLALVSIKA IGWMSSHTDF GNINGCYLNY YFFLLAAIQG ATLLLFLIIS VKYDHH RDH QRSRANGVPT SRRA UniProtKB: Solute carrier family 15 member 4 |
-Macromolecule #2: CHOLESTEROL HEMISUCCINATE
| Macromolecule | Name: CHOLESTEROL HEMISUCCINATE / type: ligand / ID: 2 / Number of copies: 12 / Formula: Y01 |
|---|---|
| Molecular weight | Theoretical: 486.726 Da |
| Chemical component information |  ChemComp-Y01: |
-Macromolecule #3: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 3 / Number of copies: 2 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #4: (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-...
| Macromolecule | Name: (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE type: ligand / ID: 4 / Number of copies: 2 / Formula: PGT |
|---|---|
| Molecular weight | Theoretical: 751.023 Da |
| Chemical component information |  ChemComp-PGT: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 6.5 / Details: 20mM MES pH6.5/ 150mM NaCl, |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER / Details: Alpha fold |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.81 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cryoSPARC / Number images used: 438976 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: cryoSPARC |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)