[English] 日本語
 Yorodumi
Yorodumi- EMDB-18616: E2/E3BP core of the human pyruvate dehydrogenase complex (map 1; ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | E2/E3BP core of the human pyruvate dehydrogenase complex (map 1; 3.4 A) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | pyruvate dehydrogenase complex / PDHc / E2 / E3BP / cryo-EM / TRANSFERASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationPDH complex synthesizes acetyl-CoA from PYR / dihydrolipoyllysine-residue acetyltransferase / dihydrolipoyllysine-residue acetyltransferase activity / Regulation of pyruvate dehydrogenase (PDH) complex / pyruvate catabolic process / Protein lipoylation / pyruvate decarboxylation to acetyl-CoA / pyruvate dehydrogenase complex / Signaling by Retinoic Acid / acyltransferase activity ...PDH complex synthesizes acetyl-CoA from PYR / dihydrolipoyllysine-residue acetyltransferase / dihydrolipoyllysine-residue acetyltransferase activity / Regulation of pyruvate dehydrogenase (PDH) complex / pyruvate catabolic process / Protein lipoylation / pyruvate decarboxylation to acetyl-CoA / pyruvate dehydrogenase complex / Signaling by Retinoic Acid / acyltransferase activity / tricarboxylic acid cycle / glucose metabolic process / mitochondrial matrix / mitochondrion / nucleoplasm / identical protein binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
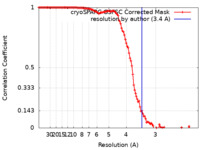
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | Zdanowicz R / Afanasyev P / Boehringer D / Glockshuber R | |||||||||
| Funding support |  Switzerland, 1 items Switzerland, 1 items
| |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2024 Journal: Sci Adv / Year: 2024Title: Stoichiometry and architecture of the human pyruvate dehydrogenase complex. Authors: Rafal Zdanowicz / Pavel Afanasyev / Adam Pruška / Julian A Harrison / Christoph Giese / Daniel Boehringer / Alexander Leitner / Renato Zenobi / Rudi Glockshuber /  Abstract: The pyruvate dehydrogenase complex (PDHc) is a key megaenzyme linking glycolysis with the citric acid cycle. In mammalian PDHc, dihydrolipoamide acetyltransferase (E2) and the dihydrolipoamide ...The pyruvate dehydrogenase complex (PDHc) is a key megaenzyme linking glycolysis with the citric acid cycle. In mammalian PDHc, dihydrolipoamide acetyltransferase (E2) and the dihydrolipoamide dehydrogenase-binding protein (E3BP) form a 60-subunit core that associates with the peripheral subunits pyruvate dehydrogenase (E1) and dihydrolipoamide dehydrogenase (E3). The structure and stoichiometry of the fully assembled, mammalian PDHc or its core remained elusive. Here, we demonstrate that the human PDHc core is formed by 48 E2 copies that bind 48 E1 heterotetramers and 12 E3BP copies that bind 12 E3 homodimers. Cryo-electron microscopy, together with native and cross-linking mass spectrometry, confirmed a core model in which 8 E2 homotrimers and 12 E2-E2-E3BP heterotrimers assemble into a pseudoicosahedral particle such that the 12 E3BP molecules form six E3BP-E3BP intertrimer interfaces distributed tetrahedrally within the 60-subunit core. The even distribution of E3 subunits in the peripheral shell of PDHc guarantees maximum enzymatic activity of the megaenzyme. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18616.map.gz emd_18616.map.gz | 13.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18616-v30.xml emd-18616-v30.xml emd-18616.xml emd-18616.xml | 18.6 KB 18.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_18616_fsc.xml emd_18616_fsc.xml | 11.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_18616.png emd_18616.png | 150.1 KB | ||
| Filedesc metadata |  emd-18616.cif.gz emd-18616.cif.gz | 5.8 KB | ||
| Others |  emd_18616_half_map_1.map.gz emd_18616_half_map_1.map.gz emd_18616_half_map_2.map.gz emd_18616_half_map_2.map.gz | 127.9 MB 127.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18616 http://ftp.pdbj.org/pub/emdb/structures/EMD-18616 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18616 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18616 | HTTPS FTP |
-Validation report
| Summary document |  emd_18616_validation.pdf.gz emd_18616_validation.pdf.gz | 760.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_18616_full_validation.pdf.gz emd_18616_full_validation.pdf.gz | 759.8 KB | Display | |
| Data in XML |  emd_18616_validation.xml.gz emd_18616_validation.xml.gz | 20.2 KB | Display | |
| Data in CIF |  emd_18616_validation.cif.gz emd_18616_validation.cif.gz | 26.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18616 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18616 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18616 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18616 | HTTPS FTP |
-Related structure data
| Related structure data |  8piuC C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_18616.map.gz / Format: CCP4 / Size: 163.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18616.map.gz / Format: CCP4 / Size: 163.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
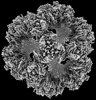
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.0929 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_18616_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_18616_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : E2/E3BP core of the human pyruvate dehydrogenase complex
| Entire | Name: E2/E3BP core of the human pyruvate dehydrogenase complex |
|---|---|
| Components |
|
-Supramolecule #1: E2/E3BP core of the human pyruvate dehydrogenase complex
| Supramolecule | Name: E2/E3BP core of the human pyruvate dehydrogenase complex type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 3.4 MDa |
-Macromolecule #1: Dihydrolipoyllysine-residue acetyltransferase component of pyruva...
| Macromolecule | Name: Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex type: protein_or_peptide / ID: 1 / Enantiomer: LEVO / EC number: dihydrolipoyllysine-residue acetyltransferase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Sequence | String: MHHHHHHENL YFQSLPPHQK VPLPSLSPTM QAGTIARWEK KEGDKINEGD LIAEVETDKA TVGFESLEEC YMAKILVAEG TRDVPIGAII CITVGKPEDI EAFKNYTLDS SAAPTPQAAP APTPAATASP PTPSAQAPGS SYPPHMQVLL PALSPTMTMG TVQRWEKKVG ...String: MHHHHHHENL YFQSLPPHQK VPLPSLSPTM QAGTIARWEK KEGDKINEGD LIAEVETDKA TVGFESLEEC YMAKILVAEG TRDVPIGAII CITVGKPEDI EAFKNYTLDS SAAPTPQAAP APTPAATASP PTPSAQAPGS SYPPHMQVLL PALSPTMTMG TVQRWEKKVG EKLSEGDLLA EIETDKATIG FEVQEEGYLA KILVPEGTRD VPLGTPLCII VEKEADISAF ADYRPTEVTD LKPQVPPPTP PPVAAVPPTP QPLAPTPSAP CPATPAGPKG RVFVSPLAKK LAVEKGIDLT QVKGTGPDGR ITKKDIDSFV PSKVAPAPAA VVPPTGPGMA PVPTGVFTDI PISNIRRVIA QRLMQSKQTI PHYYLSIDVN MGEVLLVRKE LNKILEGRSK ISVNDFIIKA SALACLKVPE ANSSWMDTVI RQNHVVDVSV AVSTPAGLIT PIVFNAHIKG VETIANDVVS LATKAREGKL QPHEFQGGTF TISNLGMFGI KNFSAIINPP QACILAIGAS EDKLVPADNE KGFDVASMMS VTLSCDHRVV DGAVGAQWLA EFRKYLEKPI TMLL UniProtKB: Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, mitochondrial |
-Macromolecule #2: Pyruvate dehydrogenase protein X component
| Macromolecule | Name: Pyruvate dehydrogenase protein X component / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Sequence | String: MGDPIKILMP SLSPTMEEGN IVKWLKKEGE AVSAGDALCE IETDKAVVTL DASDDGILAK IVVEEGSKNI RLGSLIGLIV EEGEDWKHVE IPKDVGPPPP VSKPSEPRPS PEPQISIPVK KEHIPGTLRF RLSPAARNIL EKHSLDASQG TATGPRGIFT KEDALKLVQL ...String: MGDPIKILMP SLSPTMEEGN IVKWLKKEGE AVSAGDALCE IETDKAVVTL DASDDGILAK IVVEEGSKNI RLGSLIGLIV EEGEDWKHVE IPKDVGPPPP VSKPSEPRPS PEPQISIPVK KEHIPGTLRF RLSPAARNIL EKHSLDASQG TATGPRGIFT KEDALKLVQL KQTGKITESR PTPAPTATPT APSPLQATAG PSYPRPVIPP VSTPGQPNAV GTFTEIPASN IRRVIAKRLT ESKSTVPHAY ATADCDLGAV LKVRQDLVKD DIKVSVNDFI IKAAAVTLKQ MPDVNVSWDG EGPKQLPFID ISVAVATDKG LLTPIIKDAA AKGIQEIADS VKALSKKARD GKLLPEEYQG GSFSISNLGM FGIDEFTAVI NPPQACILAV GRFRPVLKLT EDEEGNAKLQ QRQLITVTMS SDSRVVDDEL ATRFLKSFKA NLENPIRLA UniProtKB: Pyruvate dehydrogenase protein X component, mitochondrial |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Support film - Material: CARBON / Support film - topology: HOLEY |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Detector mode: COUNTING / Average electron dose: 80.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.8000000000000003 µm / Nominal defocus min: 1.6 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)