+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | SWR1-nucleosome complex in configuration 1 | ||||||||||||
 マップデータ マップデータ | |||||||||||||
 試料 試料 |
| ||||||||||||
 キーワード キーワード | Chromatin remodelling complex / nucleosome / protein-DNA complex / DNA BINDING PROTEIN | ||||||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報sexual sporulation resulting in formation of a cellular spore / cupric reductase (NADH) activity / TTT Hsp90 cochaperone complex / HATs acetylate histones / global genome nucleotide-excision repair / RNA polymerase I upstream activating factor complex / Condensation of Prophase Chromosomes / SIRT1 negatively regulates rRNA expression / Activated PKN1 stimulates transcription of AR (androgen receptor) regulated genes KLK2 and KLK3 / Assembly of the ORC complex at the origin of replication ...sexual sporulation resulting in formation of a cellular spore / cupric reductase (NADH) activity / TTT Hsp90 cochaperone complex / HATs acetylate histones / global genome nucleotide-excision repair / RNA polymerase I upstream activating factor complex / Condensation of Prophase Chromosomes / SIRT1 negatively regulates rRNA expression / Activated PKN1 stimulates transcription of AR (androgen receptor) regulated genes KLK2 and KLK3 / Assembly of the ORC complex at the origin of replication / R2TP complex / HDACs deacetylate histones / protein targeting to vacuole / Swr1 complex / Ino80 complex / Recruitment and ATM-mediated phosphorylation of repair and signaling proteins at DNA double strand breaks / Oxidative Stress Induced Senescence / RMTs methylate histone arginines / DNA damage tolerance / box C/D snoRNP assembly / SUMOylation of chromatin organization proteins / recombinational repair / 3'-5' DNA helicase activity / RNA Polymerase I Promoter Escape / NuA4 histone acetyltransferase complex / positive regulation of transcription by RNA polymerase I / nucleolar large rRNA transcription by RNA polymerase I / Estrogen-dependent gene expression / rRNA transcription / intracellular copper ion homeostasis / Ub-specific processing proteases / nucleosome binding / CENP-A containing nucleosome / nuclear periphery / DNA helicase activity / aerobic respiration / helicase activity / rRNA processing / structural constituent of chromatin / nucleosome / heterochromatin formation / nucleosome assembly / chromatin organization / 5'-3' DNA helicase activity / histone binding / molecular adaptor activity / DNA helicase / protein stabilization / chromatin remodeling / protein heterodimerization activity / DNA repair / regulation of DNA-templated transcription / regulation of transcription by RNA polymerase II / chromatin / structural molecule activity / negative regulation of transcription by RNA polymerase II / ATP hydrolysis activity / DNA binding / zinc ion binding / ATP binding / identical protein binding / nucleus / cytoplasm / cytosol 類似検索 - 分子機能 | ||||||||||||
| 生物種 |   | ||||||||||||
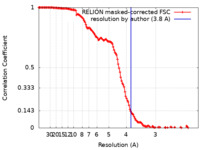
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 3.8 Å | ||||||||||||
 データ登録者 データ登録者 | Jalal ASB / Wigley DB | ||||||||||||
| 資金援助 |  英国, 3件 英国, 3件
| ||||||||||||
 引用 引用 |  ジャーナル: Nature / 年: 2024 ジャーナル: Nature / 年: 2024タイトル: Nucleosome flipping drives kinetic proofreading and processivity by SWR1. 著者: Paul Girvan / Adam S B Jalal / Elizabeth A McCormack / Michael T Skehan / Carol L Knight / Dale B Wigley / David S Rueda /  要旨: The yeast SWR1 complex catalyses the exchange of histone H2A-H2B dimers in nucleosomes, with Htz1-H2B dimers. Here we used single-molecule analysis to demonstrate two-step double exchange of the two ...The yeast SWR1 complex catalyses the exchange of histone H2A-H2B dimers in nucleosomes, with Htz1-H2B dimers. Here we used single-molecule analysis to demonstrate two-step double exchange of the two H2A-H2B dimers in a canonical yeast nucleosome with Htz1-H2B dimers, and showed that double exchange can be processive without release of the nucleosome from the SWR1 complex. Further analysis showed that bound nucleosomes flip between two states, with each presenting a different face, and hence histone dimer, to SWR1. The bound dwell time is longer when an H2A-H2B dimer is presented for exchange than when presented with an Htz1-H2B dimer. A hexasome intermediate in the reaction is bound to the SWR1 complex in a single orientation with the 'empty' site presented for dimer insertion. Cryo-electron microscopy analysis revealed different populations of complexes showing nucleosomes caught 'flipping' between different conformations without release, each placing a different dimer into position for exchange, with the Swc2 subunit having a key role in this process. Together, the data reveal a processive mechanism for double dimer exchange that explains how SWR1 can 'proofread' the dimer identities within nucleosomes. | ||||||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| 添付画像 |
|---|
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_18471.map.gz emd_18471.map.gz | 13.2 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-18471-v30.xml emd-18471-v30.xml emd-18471.xml emd-18471.xml | 34.6 KB 34.6 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| FSC (解像度算出) |  emd_18471_fsc.xml emd_18471_fsc.xml | 13.6 KB | 表示 |  FSCデータファイル FSCデータファイル |
| 画像 |  emd_18471.png emd_18471.png | 38.9 KB | ||
| Filedesc metadata |  emd-18471.cif.gz emd-18471.cif.gz | 9.9 KB | ||
| その他 |  emd_18471_half_map_1.map.gz emd_18471_half_map_1.map.gz emd_18471_half_map_2.map.gz emd_18471_half_map_2.map.gz | 171.9 MB 171.9 MB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18471 http://ftp.pdbj.org/pub/emdb/structures/EMD-18471 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18471 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18471 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_18471_validation.pdf.gz emd_18471_validation.pdf.gz | 740.3 KB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_18471_full_validation.pdf.gz emd_18471_full_validation.pdf.gz | 739.8 KB | 表示 | |
| XML形式データ |  emd_18471_validation.xml.gz emd_18471_validation.xml.gz | 21.2 KB | 表示 | |
| CIF形式データ |  emd_18471_validation.cif.gz emd_18471_validation.cif.gz | 28.1 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18471 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18471 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18471 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18471 | HTTPS FTP |
-関連構造データ
| 関連構造データ |  8qkuMC  8qkvC M: このマップから作成された原子モデル C: 同じ文献を引用 ( |
|---|---|
| 類似構造データ | 類似検索 - 機能・相同性  F&H 検索 F&H 検索 |
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| 「今月の分子」の関連する項目 |
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_18471.map.gz / 形式: CCP4 / 大きさ: 216 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_18471.map.gz / 形式: CCP4 / 大きさ: 216 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||
| 密度 |
| ||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
|
-添付データ
-ハーフマップ: #2
| ファイル | emd_18471_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 |
| ||||||||||||
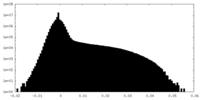
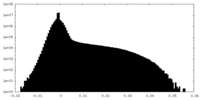
| 密度ヒストグラム |
-ハーフマップ: #1
| ファイル | emd_18471_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 |
| ||||||||||||
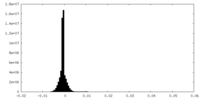
| 密度ヒストグラム |
- 試料の構成要素
試料の構成要素
+全体 : SWR1-nucleosome complex
+超分子 #1: SWR1-nucleosome complex
+分子 #1: Histone H3
+分子 #2: Histone H4
+分子 #3: Histone H2A.2
+分子 #4: Histone H2B.1
+分子 #7: Helicase SWR1
+分子 #8: Actin-like protein ARP6
+分子 #9: Vacuolar protein sorting-associated protein 71
+分子 #10: RuvB-like protein 1
+分子 #11: RuvB-like protein 2
+分子 #12: Vacuolar protein sorting-associated protein 72
+分子 #5: DNA (177-MER)
+分子 #6: DNA (177-MER)
+分子 #13: ADENOSINE-5'-DIPHOSPHATE
+分子 #14: BERYLLIUM TRIFLUORIDE ION
+分子 #15: MAGNESIUM ION
+分子 #16: ZINC ION
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 緩衝液 | pH: 7 |
|---|---|
| 凍結 | 凍結剤: ETHANE |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI TITAN KRIOS |
|---|---|
| 撮影 | フィルム・検出器のモデル: FEI FALCON IV (4k x 4k) 平均電子線量: 40.0 e/Å2 |
| 電子線 | 加速電圧: 300 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | 照射モード: SPOT SCAN / 撮影モード: BRIGHT FIELD / 最大 デフォーカス(公称値): 2.1 µm 最小 デフォーカス(公称値): 0.7000000000000001 µm |
| 実験機器 |  モデル: Titan Krios / 画像提供: FEI Company |
 ムービー
ムービー コントローラー
コントローラー
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)































 Trichoplusia ni (イラクサキンウワバ)
Trichoplusia ni (イラクサキンウワバ)