+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of SidH from Legionella pneumophila | |||||||||
 Map data Map data | Local resolution filtered EM map of SidH from Legionella pneumophila | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | SidH / Legionella / tRNA-binding / EF-Tu-binding / RNA BINDING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationguanyl-nucleotide exchange factor complex / protein-synthesizing GTPase / guanosine tetraphosphate binding / translation elongation factor activity / GTPase activity / GTP binding / magnesium ion binding / plasma membrane / cytosol Similarity search - Function | |||||||||
| Biological species |   | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.7 Å | |||||||||
 Authors Authors | Sharma R / Weis F / Bhogaraju S | |||||||||
| Funding support |  France, 1 items France, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Structural basis for the toxicity of Legionella pneumophila effector SidH. Authors: Rahul Sharma / Michael Adams / Simonne Griffith-Jones / Tobias Sahr / Laura Gomez-Valero / Felix Weis / Michael Hons / Sarah Gharbi / Rayene Berkane / Alexandra Stolz / Carmen Buchrieser / Sagar Bhogaraju /   Abstract: Legionella pneumophila (LP) secretes more than 300 effectors into the host cytosol to facilitate intracellular replication. One of these effectors, SidH, 253 kDa in size with no sequence similarity ...Legionella pneumophila (LP) secretes more than 300 effectors into the host cytosol to facilitate intracellular replication. One of these effectors, SidH, 253 kDa in size with no sequence similarity to proteins of known function is toxic when overexpressed in host cells. SidH is regulated by the LP metaeffector LubX which targets SidH for degradation in a temporal manner during LP infection. The mechanism underlying the toxicity of SidH and its role in LP infection are unknown. Here, we determined the cryo-EM structure of SidH at 2.7 Å revealing a unique alpha helical arrangement with no overall similarity to known protein structures. Surprisingly, purified SidH came bound to a E. coli EF-Tu/t-RNA/GTP ternary complex which could be modeled into the cryo-EM density. Mutation of residues disrupting the SidH-tRNA interface and SidH-EF-Tu interface abolish the toxicity of overexpressed SidH in human cells, a phenotype confirmed in infection of Acanthamoeba castellani. We also present the cryo-EM structure of SidH in complex with a U-box domain containing ubiquitin ligase LubX delineating the mechanism of regulation of SidH. Our data provide the basis for the toxicity of SidH and into its regulation by the metaeffector LubX. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18383.map.gz emd_18383.map.gz | 193.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18383-v30.xml emd-18383-v30.xml emd-18383.xml emd-18383.xml | 20.6 KB 20.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_18383_fsc.xml emd_18383_fsc.xml | 17 KB | Display |  FSC data file FSC data file |
| Images |  emd_18383.png emd_18383.png | 55.9 KB | ||
| Filedesc metadata |  emd-18383.cif.gz emd-18383.cif.gz | 7.9 KB | ||
| Others |  emd_18383_additional_1.map.gz emd_18383_additional_1.map.gz emd_18383_half_map_1.map.gz emd_18383_half_map_1.map.gz emd_18383_half_map_2.map.gz emd_18383_half_map_2.map.gz | 245.4 MB 336.8 MB 337.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18383 http://ftp.pdbj.org/pub/emdb/structures/EMD-18383 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18383 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18383 | HTTPS FTP |
-Related structure data
| Related structure data |  8qfsMC  8qhcC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_18383.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18383.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Local resolution filtered EM map of SidH from Legionella pneumophila | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.8127 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: SidH map after focussed classification on DUF domain
| File | emd_18383_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | SidH map after focussed classification on DUF domain | ||||||||||||
| Projections & Slices |
| ||||||||||||
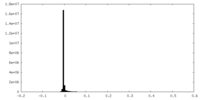
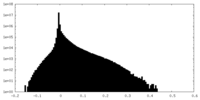
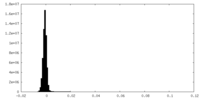
| Density Histograms |
-Half map: halfmap 1
| File | emd_18383_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | halfmap 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
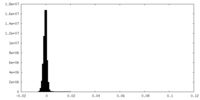
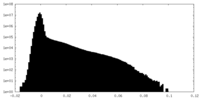
| Density Histograms |
-Half map: halfmap2
| File | emd_18383_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | halfmap2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
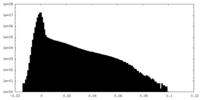
| Density Histograms |
- Sample components
Sample components
-Entire : Ternary complex of SidH with E.coli tRNA and EF-Tu
| Entire | Name: Ternary complex of SidH with E.coli tRNA and EF-Tu |
|---|---|
| Components |
|
-Supramolecule #1: Ternary complex of SidH with E.coli tRNA and EF-Tu
| Supramolecule | Name: Ternary complex of SidH with E.coli tRNA and EF-Tu / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Elongation factor Tu
| Macromolecule | Name: Elongation factor Tu / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 43.370492 KDa |
| Sequence | String: MSKEKFERTK PHVNVGTIGH VDHGKTTLTA AITTVLAKTY GGAARAFDQI DNAPEEKARG ITINTSHVEY DTPTRHYAHV DCPGHADYV KNMITGAAQM DGAILVVAAT DGPMPQTREH ILLGRQVGVP YIIVFLNKCD MVDDEELLEL VEMEVRELLS Q YDFPGDDT ...String: MSKEKFERTK PHVNVGTIGH VDHGKTTLTA AITTVLAKTY GGAARAFDQI DNAPEEKARG ITINTSHVEY DTPTRHYAHV DCPGHADYV KNMITGAAQM DGAILVVAAT DGPMPQTREH ILLGRQVGVP YIIVFLNKCD MVDDEELLEL VEMEVRELLS Q YDFPGDDT PIVRGSALKA LEGDAEWEAK ILELAGFLDS YIPEPERAID KPFLLPIEDV FSISGRGTVV TGRVERGIIK VG EEVEIVG IKETQKSTCT GVEMFRKLLD EGRAGENVGV LLRGIKREEI ERGQVLAKPG TIKPHTKFES EVYILSKDEG GRH TPFFKG YRPQFYFRTT DVTGTIELPE GVEMVMPGDN IKMVVTLIHP IAMDDGLRFA IREGGRTVGA GVVAKVLS UniProtKB: Elongation factor Tu |
-Macromolecule #3: Protein SidH
| Macromolecule | Name: Protein SidH / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 255.821562 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKHHHHHHHH HHSAGLEVLF QGPMKRTIET YIIYLKEDLK IADTCKTIKD GLLKSITDKT HFSEELATYF ERDNPNAPFK VNTTDPTQV AVLKKLLNAL ENAEKSFRAI ENIDISRDRY TAMIAKDAVM VSYKAVHEIY AALQLINHSN SDIQDIVGPH I QKLLPQMA ...String: MKHHHHHHHH HHSAGLEVLF QGPMKRTIET YIIYLKEDLK IADTCKTIKD GLLKSITDKT HFSEELATYF ERDNPNAPFK VNTTDPTQV AVLKKLLNAL ENAEKSFRAI ENIDISRDRY TAMIAKDAVM VSYKAVHEIY AALQLINHSN SDIQDIVGPH I QKLLPQMA LASKALGNFA PEHPEESAGA VLAGVVNMLP TEKPTESESL GKLSNLIFEL PHYFEELQKL IATGASGIAT KS ITSAEDY QSAMIKKANE TKYYFEQLSS KSGLLAIPSY LSIVKRLIAH STDLVNAGAP LTKQAYLDAV AKLEDIKHNI LPQ LISELE MVEESMGLKP GLLTDPALEQ MNKYYTQLAE QVDNIAKAAG VLDTVSDYSD SIGGKIVRFL AGDSKKLDVG PKLT PAPDL GVLMDDVFIQ KRRSNQESRL NEARLSSEDK SVLAAANRFF DKIGSYNSIH KAWSKWSLAN ISQSEKDALI KEYKQ FQPH FAALYPDIDK LVVDALTQPT GSNIVSRLYS SDYKQLWSSD HFKQVLSCKD SVLSSIQQSL AQSEFKAKLI EKTMSH SEE TAYSMNNKTT NLTTRVQPFE PLKFTLEDDK PVEYYHKRVI AASNQILELE RAQKGVAEFF NYIQKKYPHE NPSFDSL DE SDKEFLRKAY KTFQPQLLAL KHDDINTRLV SSLTSSKPTD PPLRLTDLVS LKSGINDYLN EKISDLNQDK TTLLDKEE E AREEQYAKNP LVAKGAELEK QTLFGQMSKL KLSKSVDDFF NKKFQTYLKD NLSPEVWKQL SSNGETLDFD KIPYLEFHK DSPEVAMYKQ LINSMHYMKN GLEKLESLND YGDPNNIYHR TRFVMTTFNA LVMNICFSKY YVMEAGNNPG LKAIVQEGLD LLKPLEGMP LIGDYLKTTE KQEPPKQNII TAWKKQQAVV ESGLSKGPKP KTDQQLISEQ LGKIQEAIDN FDGDLEVSDS A REKIKTQI GEFAKGISGL SFGPGSVKKI LAALTKLETQ LSNLDKESPE VTLGKLKDIH SELNAQFRAA AEYTEYHSGQ KF GSYSNNI STIVSNFCNG LVSNLPLKQE PAPKVKAPEK PVTPVITGTT NPHEVVFGTK HEEFNSVYQP YLLLKRITDE FRD QNNPYK PSFDELKEEA VSYYDKIQPL LESVDPKFDK NFIAKHKESS TLLKAIDEVM SMRQRINNPE SSFAKLKDLH LEGD FEKEE NKEKFRQLYK EIQPYLHKID STYDQTQFLE GLKTAKDFSG ALQRIMNEEN ALQQSTSLKD TSYLQLIAES LYQIP VKLN KLKAEPDTPE PSKEEIDANV KAFVEGLNGL SFGPGSVKKI LSTAAKLQMQ LSDIGKEGRE LTMGRLKEIQ AEFGTI LMA AADNAEFHLG LKPGTYSRTV SERFEKFYSS LIVNLPLEKD QTGLELLIDT TSTQKRLARE MERLESVKED TSAIDTK KS IFGTEHEQFS TLYQPYASLR HIAKDIEDGV HMNLYERTLE ELKEEASNEY KKIQPYLAKI NPEFTEDYIS KTDGEYSL L HAIDRVFEER HKINKPSSPF DKLRDLYLDG DFEKEENKEQ FLQLYAELQP HLIKINYQYD LAYFLRELQT PEDFKAATE RIINDESKLQ ELITGLDDTK RLKVKLCEER IGYFIDLLKK QELEVGPEKI QAFKEKIFFN YIHANVNSAL DAKIGSHAEQ FLQFIEKDF LDKKNEILEK ITIDQDMEEE IAKAIDRIAP DIINNKIESF KKLLFDSYIQ SDIKNSLHNE LGIYTSLFID K ISPEIHLH QSEILGNVAF DSKMGAEIGS KINAITPGIL LNNNPLKEAY VDLNNTLKEI NTLLDEENKK TRDNPCRNEK IA KLTSLKD RLSDLDSIPK ENTVEFLKKM QEETRSSLKS LESNDALINI YDVLNSLKET IENGSDLPEI KKDKLQMISD VQN ILSNFD KNPAERLNLA VQILNDSNPE VLSKTKGNFL IGEAFKGQVF TNYINTKISE QLNNELGPYG KVFLKQIMPD FIAK KSEII KEIAIDNMET GLETQFKIHA PAIFEKNKEL KAAYEQLNVH LKEVQSLIEA EEKKPKGNPC REEKIAALRS HQSQL MNTQ RIPDHETLRF LQEQNKSAKS FMGKLEKYDT MISVYDSLTE IREHVSNHKS LSKEIKDEKI QEISKMEDML KTTSKE PSI RLAEVKAHGL SDQCKNVLLK NSDNFLVSFF KTLFSKLFNI KNENETLVSS FKQRLQNIKG PEPVATPMET PENEAPL VN ANITRF UniProtKB: SidH |
-Macromolecule #2: tRNA
| Macromolecule | Name: tRNA / type: rna / ID: 2 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 24.620766 KDa |
| Sequence | String: GCCCGGA(4SU)AG CUCAG(H2U)CGG(H2U) AGAGCAGGGG AUUGAAAAUC CCCGUG(3AU)CCU UGG(5MU)(PSU)C GAU UCCGAGUCCG GGCACCA |
-Macromolecule #4: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 4 / Number of copies: 1 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #5: GUANOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: GUANOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 5 / Number of copies: 1 / Formula: GTP |
|---|---|
| Molecular weight | Theoretical: 523.18 Da |
| Chemical component information |  ChemComp-GTP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 41.93 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)