+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | MTHFR + SAM inhibited state | |||||||||
 Map data Map data | Main Sharpened | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Inhibited / allosteric / folate / S-Adenosylmethionin / FLAVOPROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationmethylenetetrahydrofolate reductase (NADPH) / response to vitamin B2 / methylenetetrahydrofolate reductase (NADPH) activity / methylenetetrahydrofolate reductase [NAD(P)H] activity / methionine metabolic process / modified amino acid binding / heterochromatin organization / homocysteine metabolic process / S-adenosylmethionine metabolic process / response to folic acid ...methylenetetrahydrofolate reductase (NADPH) / response to vitamin B2 / methylenetetrahydrofolate reductase (NADPH) activity / methylenetetrahydrofolate reductase [NAD(P)H] activity / methionine metabolic process / modified amino acid binding / heterochromatin organization / homocysteine metabolic process / S-adenosylmethionine metabolic process / response to folic acid / Metabolism of folate and pterines / : / tetrahydrofolate interconversion / response to amino acid / FAD binding / response to interleukin-1 / neural tube closure / NADP binding / flavin adenine dinucleotide binding / response to hypoxia / response to xenobiotic stimulus / protein-containing complex binding / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
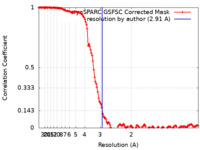
| Method | single particle reconstruction / cryo EM / Resolution: 2.91 Å | |||||||||
 Authors Authors | Blomgren LKM / Yue WW / Froese DS / McCorvie TJ | |||||||||
| Funding support |  Switzerland, 1 items Switzerland, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Dynamic inter-domain transformations mediate the allosteric regulation of human 5, 10-methylenetetrahydrofolate reductase. Authors: Linnea K M Blomgren / Melanie Huber / Sabrina R Mackinnon / Céline Bürer / Arnaud Baslé / Wyatt W Yue / D Sean Froese / Thomas J McCorvie /   Abstract: 5,10-methylenetetrahydrofolate reductase (MTHFR) commits folate-derived one-carbon units to generate the methyl-donor S-adenosyl-L-methionine (SAM). Eukaryotic MTHFR appends to the well-conserved ...5,10-methylenetetrahydrofolate reductase (MTHFR) commits folate-derived one-carbon units to generate the methyl-donor S-adenosyl-L-methionine (SAM). Eukaryotic MTHFR appends to the well-conserved catalytic domain (CD) a unique regulatory domain (RD) that confers feedback inhibition by SAM. Here we determine the cryo-electron microscopy structures of human MTHFR bound to SAM and its demethylated product S-adenosyl-L-homocysteine (SAH). In the active state, with the RD bound to a single SAH, the CD is flexible and exposes its active site for catalysis. However, in the inhibited state the RD pocket is remodelled, exposing a second SAM-binding site that was previously occluded. Dual-SAM bound MTHFR demonstrates a substantially rearranged inter-domain linker that reorients the CD, inserts a loop into the active site, positions Tyr404 to bind the cofactor FAD, and blocks substrate access. Our data therefore explain the long-distance regulatory mechanism of MTHFR inhibition, underpinned by the transition between dual-SAM and single-SAH binding in response to cellular methylation status. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18300.map.gz emd_18300.map.gz | 229.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18300-v30.xml emd-18300-v30.xml emd-18300.xml emd-18300.xml | 20.9 KB 20.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_18300_fsc.xml emd_18300_fsc.xml | 13.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_18300.png emd_18300.png | 62.9 KB | ||
| Masks |  emd_18300_msk_1.map emd_18300_msk_1.map | 244.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-18300.cif.gz emd-18300.cif.gz | 6.5 KB | ||
| Others |  emd_18300_additional_1.map.gz emd_18300_additional_1.map.gz emd_18300_half_map_1.map.gz emd_18300_half_map_1.map.gz emd_18300_half_map_2.map.gz emd_18300_half_map_2.map.gz | 120.7 MB 226.2 MB 226.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18300 http://ftp.pdbj.org/pub/emdb/structures/EMD-18300 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18300 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18300 | HTTPS FTP |
-Validation report
| Summary document |  emd_18300_validation.pdf.gz emd_18300_validation.pdf.gz | 1.2 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_18300_full_validation.pdf.gz emd_18300_full_validation.pdf.gz | 1.2 MB | Display | |
| Data in XML |  emd_18300_validation.xml.gz emd_18300_validation.xml.gz | 22.3 KB | Display | |
| Data in CIF |  emd_18300_validation.cif.gz emd_18300_validation.cif.gz | 28.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18300 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18300 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18300 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18300 | HTTPS FTP |
-Related structure data
| Related structure data |  8qa6MC  8qa4C  8qa5C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_18300.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18300.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Main Sharpened | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.574 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_18300_msk_1.map emd_18300_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Non-sharpened
| File | emd_18300_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Non-sharpened | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_18300_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_18300_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Human 5,10-methylenetetrahydrofolate reductase in complex with S-...
| Entire | Name: Human 5,10-methylenetetrahydrofolate reductase in complex with S-Adenosyl-L-Methionine |
|---|---|
| Components |
|
-Supramolecule #1: Human 5,10-methylenetetrahydrofolate reductase in complex with S-...
| Supramolecule | Name: Human 5,10-methylenetetrahydrofolate reductase in complex with S-Adenosyl-L-Methionine type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 150 KDa |
-Macromolecule #1: Methylenetetrahydrofolate reductase (NADPH)
| Macromolecule | Name: Methylenetetrahydrofolate reductase (NADPH) / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 75.461195 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MVNEARGNSS LNPCLEGSAS SGSESSKDSS RCSTPGLDPE RHERLREKMR RRLESGDKWF SLEFFPPRTA EGAVNLISRF DRMAAGGPL YIDVTWHPAG DPGSDKETSS MMIASTAVNY CGLETILHMT CCRQRLEEIT GHLHKAKQLG LKNIMALRGD P IGDQWEEE ...String: MVNEARGNSS LNPCLEGSAS SGSESSKDSS RCSTPGLDPE RHERLREKMR RRLESGDKWF SLEFFPPRTA EGAVNLISRF DRMAAGGPL YIDVTWHPAG DPGSDKETSS MMIASTAVNY CGLETILHMT CCRQRLEEIT GHLHKAKQLG LKNIMALRGD P IGDQWEEE EGGFNYAVDL VKHIRSEFGD YFDICVAGYP KGHPEAGSFE ADLKHLKEKV SAGADFIITQ LFFEADTFFR FV KACTDMG ITCPIVPGIF PIQGYHSLRQ LVKLSKLEVP QEIKDVIEPI KDNDAAIRNY GIELAVSLCQ ELLASGLVPG LHF YTLNRE MATTEVLKRL GMWTEDPRRP LPWALSAHPK RREEDVRPIF WASRPKSYIY RTQEWDEFPN GRWGNSSSPA FGEL KDYYL FYLKSKSPKE ELLKMWGEEL TSEASVFEVF VLYLSGEPNR NGHKVTCLPW NDEPLAAETS LLKEELLRVN RQGIL TINS QPNINGKPSS DPIVGWGPSG GYVFQKAYLE FFTSRETAEA LLQVLKKYEL RVNYHLVNVK GENITNAPEL QPNAVT WGI FPGREIIQPT VVDPVSFMFW KDEAFALWIE QWGKLYEEES PSRTIIQYIH DNYFLVNLVD NDFPLDNCLW QVVEDTL EL LNRPTQNARE TEAPAENLYF Q UniProtKB: Methylenetetrahydrofolate reductase (NADPH) |
-Macromolecule #2: S-ADENOSYLMETHIONINE
| Macromolecule | Name: S-ADENOSYLMETHIONINE / type: ligand / ID: 2 / Number of copies: 4 / Formula: SAM |
|---|---|
| Molecular weight | Theoretical: 398.437 Da |
| Chemical component information |  ChemComp-SAM: |
-Macromolecule #3: FLAVIN-ADENINE DINUCLEOTIDE
| Macromolecule | Name: FLAVIN-ADENINE DINUCLEOTIDE / type: ligand / ID: 3 / Number of copies: 2 / Formula: FAD |
|---|---|
| Molecular weight | Theoretical: 785.55 Da |
| Chemical component information |  ChemComp-FAD: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.0 mg/mL |
|---|---|
| Buffer | pH: 7.5 Details: 20 mM HEPES, pH 7.5, 150 mM NaCl, 5 mM S-Adenosyl-L-Methionine, filter sterilised |
| Grid | Model: Au-flat 1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: GOLD / Support film - topology: HOLEY ARRAY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 295 K / Instrument: FEI VITROBOT MARK III |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Number grids imaged: 3 / Number real images: 2394 / Average exposure time: 6.17 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.1 µm / Nominal defocus min: 0.9 µm / Nominal magnification: 240000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Overall B value: 105.2 / Target criteria: Cross-correlation coeficient | |||||||||
| Output model |  PDB-8qa6: |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)