[English] 日本語
 Yorodumi
Yorodumi- EMDB-18235: Structure of the recycling U5 snRNP bound to chaperone CD2BP2 (St... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the recycling U5 snRNP bound to chaperone CD2BP2 (State 3, Map 3) | |||||||||
 Map data Map data | sharpened map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | U5 snRNP / CD2BP2 / TSSC4 / spliceosome / splicing | |||||||||
| Function / homology |  Function and homology information Function and homology informationRNA localization / R-loop processing / U2 snRNP binding / U7 snRNA binding / histone pre-mRNA DCP binding / U7 snRNP / cis assembly of pre-catalytic spliceosome / histone pre-mRNA 3'end processing complex / SLBP independent Processing of Histone Pre-mRNAs / SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs ...RNA localization / R-loop processing / U2 snRNP binding / U7 snRNA binding / histone pre-mRNA DCP binding / U7 snRNP / cis assembly of pre-catalytic spliceosome / histone pre-mRNA 3'end processing complex / SLBP independent Processing of Histone Pre-mRNAs / SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs / spliceosome conformational change to release U4 (or U4atac) and U1 (or U11) / U12-type spliceosomal complex / protein methylation / 7-methylguanosine cap hypermethylation / U1 snRNP binding / U2-type catalytic step 1 spliceosome / RNA splicing, via transesterification reactions / methylosome / pICln-Sm protein complex / snRNP binding / sno(s)RNA-containing ribonucleoprotein complex / small nuclear ribonucleoprotein complex / SMN-Sm protein complex / spliceosomal tri-snRNP complex / U2-type precatalytic spliceosome / P granule / commitment complex / telomerase holoenzyme complex / U2-type prespliceosome assembly / U2-type spliceosomal complex / telomerase RNA binding / U2-type catalytic step 2 spliceosome / U2 snRNP / U1 snRNP / RNA Polymerase II Transcription Termination / U4 snRNP / U2-type prespliceosome / K63-linked polyubiquitin modification-dependent protein binding / precatalytic spliceosome / spliceosomal complex assembly / mRNA Splicing - Minor Pathway / spliceosomal tri-snRNP complex assembly / U5 snRNA binding / U5 snRNP / U2 snRNA binding / U6 snRNA binding / pre-mRNA intronic binding / spliceosomal snRNP assembly / ribonucleoprotein complex binding / U1 snRNA binding / RNA processing / Cajal body / U4/U6 x U5 tri-snRNP complex / catalytic step 2 spliceosome / mRNA Splicing - Major Pathway / RNA splicing / response to cocaine / spliceosomal complex / helicase activity / mRNA splicing, via spliceosome / cellular response to xenobiotic stimulus / fibrillar center / mRNA processing / osteoblast differentiation / cellular response to tumor necrosis factor / cellular response to lipopolysaccharide / snRNP Assembly / protein-macromolecule adaptor activity / SARS-CoV-2 modulates host translation machinery / RNA helicase activity / cilium / nuclear speck / nuclear body / ciliary basal body / RNA helicase / intracellular membrane-bounded organelle / GTPase activity / mRNA binding / centrosome / GTP binding / chromatin / nucleolus / enzyme binding / positive regulation of transcription by RNA polymerase II / ATP hydrolysis activity / RNA binding / extracellular exosome / nucleoplasm / ATP binding / identical protein binding / membrane / nucleus / plasma membrane / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.9 Å | |||||||||
 Authors Authors | Riabov Bassat D / Plaschka C / Vorlaender MK | |||||||||
| Funding support | European Union, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2024 Journal: Nat Struct Mol Biol / Year: 2024Title: Structural basis of human U5 snRNP late biogenesis and recycling. Authors: Daria Riabov Bassat / Supapat Visanpattanasin / Matthias K Vorländer / Laura Fin / Alexander W Phillips / Clemens Plaschka /  Abstract: Pre-mRNA splicing by the spliceosome requires the biogenesis and recycling of its small nuclear ribonucleoprotein (snRNP) complexes, which are consumed in each round of splicing. The human U5 snRNP ...Pre-mRNA splicing by the spliceosome requires the biogenesis and recycling of its small nuclear ribonucleoprotein (snRNP) complexes, which are consumed in each round of splicing. The human U5 snRNP is the ~1 MDa 'heart' of the spliceosome and is recycled through an unknown mechanism involving major architectural rearrangements and the dedicated chaperones CD2BP2 and TSSC4. Late steps in U5 snRNP biogenesis similarly involve these chaperones. Here we report cryo-electron microscopy structures of four human U5 snRNP-CD2BP2-TSSC4 complexes, revealing how a series of molecular events primes the U5 snRNP to generate the ~2 MDa U4/U6.U5 tri-snRNP, the largest building block of the spliceosome. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18235.map.gz emd_18235.map.gz | 168.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18235-v30.xml emd-18235-v30.xml emd-18235.xml emd-18235.xml | 37.6 KB 37.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_18235_fsc.xml emd_18235_fsc.xml | 11.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_18235.png emd_18235.png | 69.2 KB | ||
| Filedesc metadata |  emd-18235.cif.gz emd-18235.cif.gz | 12.2 KB | ||
| Others |  emd_18235_additional_1.map.gz emd_18235_additional_1.map.gz emd_18235_half_map_1.map.gz emd_18235_half_map_1.map.gz emd_18235_half_map_2.map.gz emd_18235_half_map_2.map.gz | 89.5 MB 165.1 MB 165.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18235 http://ftp.pdbj.org/pub/emdb/structures/EMD-18235 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18235 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18235 | HTTPS FTP |
-Related structure data
| Related structure data |  8q7wMC  8q7qC  8q7vC  8q7xC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_18235.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18235.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpened map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.24 Å | ||||||||||||||||||||||||||||||||||||
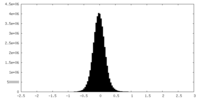
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: unsharpened map
| File | emd_18235_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
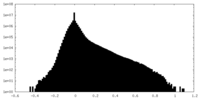
| Density Histograms |
-Half map: half map 2
| File | emd_18235_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
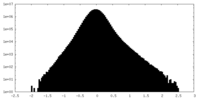
| Density Histograms |
-Half map: half map 1
| File | emd_18235_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
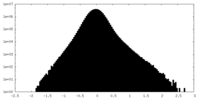
| Density Histograms |
- Sample components
Sample components
+Entire : Structure of the recycling U5 snRNP bound to chaperone CD2BP2 (St...
+Supramolecule #1: Structure of the recycling U5 snRNP bound to chaperone CD2BP2 (St...
+Macromolecule #1: U5 snRNA
+Macromolecule #2: Pre-mRNA-processing-splicing factor 8
+Macromolecule #3: U5 small nuclear ribonucleoprotein 200 kDa helicase
+Macromolecule #4: 116 kDa U5 small nuclear ribonucleoprotein component
+Macromolecule #5: U5 small nuclear ribonucleoprotein 40 kDa protein
+Macromolecule #6: Probable ATP-dependent RNA helicase DDX23
+Macromolecule #7: Pre-mRNA-processing factor 6
+Macromolecule #8: CD2 antigen cytoplasmic tail-binding protein 2
+Macromolecule #9: Small nuclear ribonucleoprotein Sm D1
+Macromolecule #10: Small nuclear ribonucleoprotein-associated proteins B and B'
+Macromolecule #11: Small nuclear ribonucleoprotein Sm D2
+Macromolecule #12: Small nuclear ribonucleoprotein Sm D3
+Macromolecule #13: Small nuclear ribonucleoprotein E
+Macromolecule #14: Small nuclear ribonucleoprotein F
+Macromolecule #15: Small nuclear ribonucleoprotein G
+Macromolecule #16: MAGNESIUM ION
+Macromolecule #17: GUANOSINE-5'-TRIPHOSPHATE
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.9 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)