+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1822 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Yeast Anaphase Promoting Complex (Dimer) | |||||||||
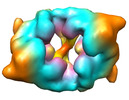
 Map data Map data | 3D structure of the Yeast Anaphase Promoting Complex (Dimer) | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Anaphase / Yeast / Cell cycle | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / negative staining / Resolution: 35.0 Å | |||||||||
 Authors Authors | Buschhorn BA / Petzold G / Galova M / Dube P / Kraft C / Herzog F / Peters JM / Stark H | |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2011 Journal: Nat Struct Mol Biol / Year: 2011Title: Substrate binding on the APC/C occurs between the coactivator Cdh1 and the processivity factor Doc1. Authors: Bettina A Buschhorn / Georg Petzold / Marta Galova / Prakash Dube / Claudine Kraft / Franz Herzog / Holger Stark / Jan-Michael Peters /   Abstract: The anaphase-promoting complex/cyclosome (APC/C) is a 22S ubiquitin ligase complex that initiates chromosome segregation and mitotic exit. We have used biochemical and electron microscopic analyses ...The anaphase-promoting complex/cyclosome (APC/C) is a 22S ubiquitin ligase complex that initiates chromosome segregation and mitotic exit. We have used biochemical and electron microscopic analyses of Saccharomyces cerevisiae and human APC/C to address how the APC/C subunit Doc1 contributes to recruitment and processive ubiquitylation of APC/C substrates, and to understand how APC/C monomers interact to form a 36S dimeric form. We show that Doc1 interacts with Cdc27, Cdc16 and Apc1 and is located in the vicinity of the cullin-RING module Apc2-Apc11 in the inner cavity of the APC/C. Substrate proteins also bind in the inner cavity, in close proximity to Doc1 and the coactivator Cdh1, and induce conformational changes in Apc2-Apc11. Our results suggest that substrates are recruited to the APC/C by binding to a bipartite substrate receptor composed of a coactivator protein and Doc1. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1822.map.gz emd_1822.map.gz | 941.9 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1822-v30.xml emd-1822-v30.xml emd-1822.xml emd-1822.xml | 6.8 KB 6.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_1822.png emd_1822.png | 145.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1822 http://ftp.pdbj.org/pub/emdb/structures/EMD-1822 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1822 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1822 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_1822.map.gz / Format: CCP4 / Size: 1001 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1822.map.gz / Format: CCP4 / Size: 1001 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 3D structure of the Yeast Anaphase Promoting Complex (Dimer) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 7.2 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Yeast anaphase promoting complex (Dimer)
| Entire | Name: Yeast anaphase promoting complex (Dimer) |
|---|---|
| Components |
|
-Supramolecule #1000: Yeast anaphase promoting complex (Dimer)
| Supramolecule | Name: Yeast anaphase promoting complex (Dimer) / type: sample / ID: 1000 / Number unique components: 1 |
|---|
-Supramolecule #1: APCC
| Supramolecule | Name: APCC / type: organelle_or_cellular_component / ID: 1 / Name.synonym: APCC / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | negative staining, cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Staining | Type: NEGATIVE / Details: Staining with 1% uranyl formate |
|---|---|
| Vitrification | Cryogen name: NITROGEN / Instrument: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | FEI/PHILIPS CM200FEG |
|---|---|
| Image recording | Digitization - Sampling interval: 1.8 µm |
| Electron beam | Acceleration voltage: 160 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD |
| Sample stage | Specimen holder: Eucentric / Specimen holder model: GATAN LIQUID NITROGEN |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Point group: C2 (2 fold cyclic) / Resolution.type: BY AUTHOR / Resolution: 35.0 Å / Resolution method: FSC 0.5 CUT-OFF / Number images used: 1271 |
|---|
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















