[English] 日本語
 Yorodumi
Yorodumi- EMDB-18190: Cryo-EM map of the trastuzumab-HER2 complex obtained from local r... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM map of the trastuzumab-HER2 complex obtained from local refinement of the HER2-pertuzumab-trastuzumab ternary complex | |||||||||
 Map data Map data | Cryo-EM map of the trastuzumab-HER2 interface obtained my local refinement of the ternary complex HER2-pertuzumab-trastuzumab Fabs | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ErbB-2 / Trastuzumab / Ternary complex / Protein / Flexibility / Continuous conformation / Cryo-EM / Single particle analysis / Structural protein | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
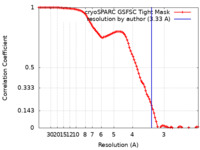
| Method | single particle reconstruction / cryo EM / Resolution: 3.33 Å | |||||||||
 Authors Authors | Ruedas R / Bressanelli S | |||||||||
| Funding support |  France, 2 items France, 2 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Atomic structure and conformational variability of the HER2-Trastuzumab-Pertuzumab complex Authors: Ruedas R / Vuillemot R / Tubiana T / Winter JM / Pieri L / Arteni AA / Samson C / Jonic J / Mathieu M / Bressanelli S | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18190.map.gz emd_18190.map.gz | 42.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18190-v30.xml emd-18190-v30.xml emd-18190.xml emd-18190.xml | 20.2 KB 20.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_18190_fsc.xml emd_18190_fsc.xml | 9.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_18190.png emd_18190.png | 116 KB | ||
| Others |  emd_18190_half_map_1.map.gz emd_18190_half_map_1.map.gz emd_18190_half_map_2.map.gz emd_18190_half_map_2.map.gz | 77.8 MB 77.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18190 http://ftp.pdbj.org/pub/emdb/structures/EMD-18190 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18190 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18190 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_18190.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18190.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM map of the trastuzumab-HER2 interface obtained my local refinement of the ternary complex HER2-pertuzumab-trastuzumab Fabs | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.16 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half map A
| File | emd_18190_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
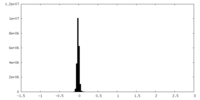
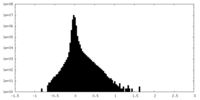
| Density Histograms |
-Half map: Half map B
| File | emd_18190_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
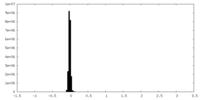
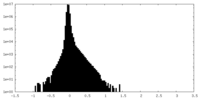
| Density Histograms |
- Sample components
Sample components
-Entire : Ternary complex of her2-trastuzumab-pertuzumab Fabs
| Entire | Name: Ternary complex of her2-trastuzumab-pertuzumab Fabs |
|---|---|
| Components |
|
-Supramolecule #1: Ternary complex of her2-trastuzumab-pertuzumab Fabs
| Supramolecule | Name: Ternary complex of her2-trastuzumab-pertuzumab Fabs / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: Fab fragment from trastuzumab and pertuzumab linked to the extracellular domain of erbb2 (her2) |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 171.364 KDa |
-Macromolecule #1: Receptor tyrosine-protein kinase HER2
| Macromolecule | Name: Receptor tyrosine-protein kinase HER2 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO / EC number: receptor protein-tyrosine kinase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: TQVCTGTDMK LRLPASPETH LDMLRHLYQG CQVVQGNLEL TYLPTNASLS FLQDIQEVQG YVLIAHNQVR QVPLQRLRIV RGTQLFEDN YALAVLDNGD PLNNTTPVTG ASPGGLRELQ LRSLTEILKG GVLIQRNPQL CYQDTILWKD IFHKNNQLAL T LIDTNRSR ...String: TQVCTGTDMK LRLPASPETH LDMLRHLYQG CQVVQGNLEL TYLPTNASLS FLQDIQEVQG YVLIAHNQVR QVPLQRLRIV RGTQLFEDN YALAVLDNGD PLNNTTPVTG ASPGGLRELQ LRSLTEILKG GVLIQRNPQL CYQDTILWKD IFHKNNQLAL T LIDTNRSR ACHPCSPMCK GSRCWGESSE DCQSLTRTVC AGGCARCKGP LPTDCCHEQC AAGCTGPKHS DCLACLHFNH SG ICELHCP ALVTYNTDTF ESMPNPEGRY TFGASCVTAC PYNYLSTDVG SCTLVCPLHN QEVTAEDGTQ RCEKCSKPCA RVC YGLGME HLREVRAVTS ANIQEFAGCK KIFGSLAFLP ESFDGDPASN TAPLQPEQLQ VFETLEEITG YLYISAWPDS LPDL SVFQN LQVIRGRILH NGAYSLTLQG LGISWLGLRS LRELGSGLAL IHHNTHLCFV HTVPWDQLFR NPHQALLHTA NRPED ECVG EGLACHQLCA RGHCWGPGPT QCVNCSQFLR GQECVEECRV LQGLPREYVN ARHCLPCHPE CQPQNGSVTC FGPEAD QCV ACAHYKDPPF CVARCPSGVK PDLSYMPIWK FPDEEGACQP CPINCTHSCV DLDDKGCPAE Q(NAG)(NAG)(NAG)(BMA)(NAG)(NAG)(BMA)(NAG)(NAG) |
-Macromolecule #2: Trastuzumab Fab heavy chain
| Macromolecule | Name: Trastuzumab Fab heavy chain / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Sequence | String: EVQLVESGGG LVQPGGSLRL SCAASGFTFT DYTMDWVRQA PGKGLEWVAD VNPNSGGSI YNQRFKGRFT LSVDRSKNTL YLQMNSLRAE DTAVYYCARN L GPSFYFDY WGQGTLVTVS SASTKGPSVF PLAPSSKSTS GGTAALGCLV KD YFPEPVT VSWNSGALTS ...String: EVQLVESGGG LVQPGGSLRL SCAASGFTFT DYTMDWVRQA PGKGLEWVAD VNPNSGGSI YNQRFKGRFT LSVDRSKNTL YLQMNSLRAE DTAVYYCARN L GPSFYFDY WGQGTLVTVS SASTKGPSVF PLAPSSKSTS GGTAALGCLV KD YFPEPVT VSWNSGALTS GVHTFPAVLQ SSGLYSLSSV VTVPSSSLGT QTY ICNVNH KPSNTKVDKK VEPKSC |
-Macromolecule #3: Trastuzumab Fab light chain
| Macromolecule | Name: Trastuzumab Fab light chain / type: protein_or_peptide / ID: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Sequence | String: DIQMTQSPSS LSASVGDRVT ITCRASQDVN TAVAWYQQKP GKAPKLLIYS ASFLYSGVP SRFSGSRSGT DFTLTISSLQ PEDFATYYCQ QHYTTPPTFG Q GTKVEIKR TVAAPSVFIF PPSDEQLKSG TASVVCLLNN FYPREAKVQW KV DNALQSG NSQESVTEQD ...String: DIQMTQSPSS LSASVGDRVT ITCRASQDVN TAVAWYQQKP GKAPKLLIYS ASFLYSGVP SRFSGSRSGT DFTLTISSLQ PEDFATYYCQ QHYTTPPTFG Q GTKVEIKR TVAAPSVFIF PPSDEQLKSG TASVVCLLNN FYPREAKVQW KV DNALQSG NSQESVTEQD SKDSTYSLSS TLTLSKADYE KHKVYACEVT HQG LSSPVT KSFNRGEC |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.12 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 288.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Number grids imaged: 1 / Number real images: 8928 / Average exposure time: 4.71 sec. / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.1 mm / Nominal defocus max: 2.6 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 240000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: HELIUM |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)