+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human KMN network (outer kinetochore) | |||||||||
 Map data Map data | Sharpened map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | OUTER KINETOCHORE / KMN / COMPLEX / CELL CYCLE | |||||||||
| Function / homology |  Function and homology information Function and homology informationKnl1/Spc105 complex / positive regulation of meiosis I spindle assembly checkpoint / homologous chromosome orientation in meiotic metaphase I / MIS12/MIND type complex / skeletal muscle satellite cell proliferation / Ndc80 complex / leucine zipper domain binding / acrosome assembly / regulation of mitotic cell cycle spindle assembly checkpoint / attachment of spindle microtubules to kinetochore ...Knl1/Spc105 complex / positive regulation of meiosis I spindle assembly checkpoint / homologous chromosome orientation in meiotic metaphase I / MIS12/MIND type complex / skeletal muscle satellite cell proliferation / Ndc80 complex / leucine zipper domain binding / acrosome assembly / regulation of mitotic cell cycle spindle assembly checkpoint / attachment of spindle microtubules to kinetochore / kinetochore assembly / outer kinetochore / attachment of mitotic spindle microtubules to kinetochore / protein localization to kinetochore / mitotic spindle assembly checkpoint signaling / mitotic sister chromatid segregation / Amplification of signal from unattached kinetochores via a MAD2 inhibitory signal / Deposition of new CENPA-containing nucleosomes at the centromere / Mitotic Prometaphase / acrosomal vesicle / EML4 and NUDC in mitotic spindle formation / Resolution of Sister Chromatid Cohesion / mitotic spindle organization / chromosome segregation / RHO GTPases Activate Formins / kinetochore / fibrillar center / spindle pole / azurophil granule lumen / Separation of Sister Chromatids / transcription regulator complex / microtubule binding / transcription by RNA polymerase II / transcription coactivator activity / nuclear speck / nuclear body / cell division / intracellular membrane-bounded organelle / Neutrophil degranulation / nucleolus / Golgi apparatus / extracellular region / nucleoplasm / nucleus / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
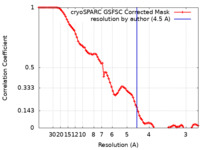
| Method | single particle reconstruction / cryo EM / Resolution: 4.5 Å | |||||||||
 Authors Authors | Raisch T / Polley S / Vetter I / Musacchio A / Raunser S | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2024 Journal: Nat Struct Mol Biol / Year: 2024Title: Structure of the human KMN complex and implications for regulation of its assembly. Authors: Soumitra Polley / Tobias Raisch / Sabrina Ghetti / Marie Körner / Melina Terbeck / Frauke Gräter / Stefan Raunser / Camilo Aponte-Santamaría / Ingrid R Vetter / Andrea Musacchio /  Abstract: Biorientation of chromosomes during cell division is necessary for precise dispatching of a mother cell's chromosomes into its two daughters. Kinetochores, large layered structures built on ...Biorientation of chromosomes during cell division is necessary for precise dispatching of a mother cell's chromosomes into its two daughters. Kinetochores, large layered structures built on specialized chromosome loci named centromeres, promote biorientation by binding and sensing spindle microtubules. One of the outer layer main components is a ten-subunit assembly comprising Knl1C, Mis12C and Ndc80C (KMN) subcomplexes. The KMN is highly elongated and docks on kinetochores and microtubules through interfaces at its opposite extremes. Here, we combine cryogenic electron microscopy reconstructions and AlphaFold2 predictions to generate a model of the human KMN that reveals all intra-KMN interfaces. We identify and functionally validate two interaction interfaces that link Mis12C to Ndc80C and Knl1C. Through targeted interference experiments, we demonstrate that this mutual organization strongly stabilizes the KMN assembly. Our work thus reports a comprehensive structural and functional analysis of this part of the kinetochore microtubule-binding machinery and elucidates the path of connections from the chromatin-bound components to the force-generating components. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18179.map.gz emd_18179.map.gz | 59.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18179-v30.xml emd-18179-v30.xml emd-18179.xml emd-18179.xml | 28.4 KB 28.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_18179_fsc.xml emd_18179_fsc.xml | 8.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_18179.png emd_18179.png | 72.1 KB | ||
| Masks |  emd_18179_msk_1.map emd_18179_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-18179.cif.gz emd-18179.cif.gz | 7.4 KB | ||
| Others |  emd_18179_additional_1.map.gz emd_18179_additional_1.map.gz emd_18179_additional_2.map.gz emd_18179_additional_2.map.gz emd_18179_half_map_1.map.gz emd_18179_half_map_1.map.gz emd_18179_half_map_2.map.gz emd_18179_half_map_2.map.gz | 32.1 MB 57.2 MB 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18179 http://ftp.pdbj.org/pub/emdb/structures/EMD-18179 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18179 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18179 | HTTPS FTP |
-Related structure data
| Related structure data |  8q5hMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_18179.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18179.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.36 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_18179_msk_1.map emd_18179_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
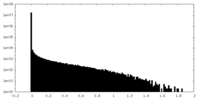
| Density Histograms |
-Additional map: Non-sharpened map
| File | emd_18179_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Non-sharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
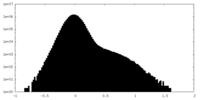
| Density Histograms |
-Additional map: Map sharpened by DeepEMhancer
| File | emd_18179_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map sharpened by DeepEMhancer | ||||||||||||
| Projections & Slices |
| ||||||||||||
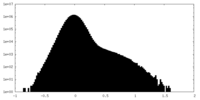
| Density Histograms |
-Half map: Half map B
| File | emd_18179_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map A
| File | emd_18179_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : KMN network
| Entire | Name: KMN network |
|---|---|
| Components |
|
-Supramolecule #1: KMN network
| Supramolecule | Name: KMN network / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Kinetochore protein Spc24
| Macromolecule | Name: Kinetochore protein Spc24 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 10.344515 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MELKEIEADL ERQEKEVDED TTVTIPSAVY VAQLYHQVSK IEWDYECEPG MVKGIHHGPS VAQPIHLDST QLSRKFISDY LWSLVDTEW UniProtKB: Kinetochore protein Spc24 |
-Macromolecule #2: Kinetochore protein Spc25
| Macromolecule | Name: Kinetochore protein Spc25 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 16.285332 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKQELEVLTA NIQDLKEEYS RKKETISTAN KANAERLKRL QKSADLYKDR LGLEIRKIYG EKLQFIFTNI DPKNPESPFM FSLHLNEAR DYEVSDSAPH LEGLAEFQEN VRKTNNFSAF LANVRKAFTA TVYNHHHHHH UniProtKB: Kinetochore protein Spc25 |
-Macromolecule #3: Protein MIS12 homolog
| Macromolecule | Name: Protein MIS12 homolog / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 24.170922 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MSVDPMTYEA QFFGFTPQTC MLRIYIAFQD YLFEVMQAVE QVILKKLDGI PDCDISPVQI RKCTEKFLCF MKGHFDNLFS KMEQLFLQL ILRIPSNILL PEDKCKETPY SEEDFQHLQK EIEQLQEKYK TELCTKQALL AELEEQKIVQ AKLKQTLTFF D ELHNVGRD ...String: MSVDPMTYEA QFFGFTPQTC MLRIYIAFQD YLFEVMQAVE QVILKKLDGI PDCDISPVQI RKCTEKFLCF MKGHFDNLFS KMEQLFLQL ILRIPSNILL PEDKCKETPY SEEDFQHLQK EIEQLQEKYK TELCTKQALL AELEEQKIVQ AKLKQTLTFF D ELHNVGRD HGTSDFRESL VSLVQNSRKL QNIRDNVEKE SKRLKIS UniProtKB: Protein MIS12 homolog |
-Macromolecule #4: Polyamine-modulated factor 1
| Macromolecule | Name: Polyamine-modulated factor 1 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.368311 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MAEASSANLG SGCEEKRHEG SSSESVPPGT TISRVKLLDT MVDTFLQKLV AAGSYQRFTD CYKCFYQLQP AMTQQIYDKF IAQLQTSIR EEISDIKEEG NLEAVLNALD KIVEEGKVRK EPAWRPSGIP EKDLHSVMAP YFLQQRDTLR RHVQKQEAEN Q QLADAVLA ...String: MAEASSANLG SGCEEKRHEG SSSESVPPGT TISRVKLLDT MVDTFLQKLV AAGSYQRFTD CYKCFYQLQP AMTQQIYDKF IAQLQTSIR EEISDIKEEG NLEAVLNALD KIVEEGKVRK EPAWRPSGIP EKDLHSVMAP YFLQQRDTLR RHVQKQEAEN Q QLADAVLA GRRQVEELQL QVQAQQQAWQ ALHREQRELV AVLREPE UniProtKB: Polyamine-modulated factor 1 |
-Macromolecule #5: Kinetochore-associated protein DSN1 homolog
| Macromolecule | Name: Kinetochore-associated protein DSN1 homolog / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 40.951062 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MTSVTRSEII DEKGPVMSKT HDHQLESSLS PVEVFAKTSA SLEMNQGVSE ERIHLGSSPK KGGNCDLSHQ ERLQSKSLHL SPQEQSASY QDRRQSWRRA SMKETNRRKS LHPIHQGITE LSRSISVDLA ESKRLGCLLL SSFQFSIQKL EPFLRDTKGF S LESFRAKA ...String: MTSVTRSEII DEKGPVMSKT HDHQLESSLS PVEVFAKTSA SLEMNQGVSE ERIHLGSSPK KGGNCDLSHQ ERLQSKSLHL SPQEQSASY QDRRQSWRRA SMKETNRRKS LHPIHQGITE LSRSISVDLA ESKRLGCLLL SSFQFSIQKL EPFLRDTKGF S LESFRAKA SSLSEELKHF ADGLETDGTL QKCFEDSNGK ASDFSLEASV AEMKEYITKF SLERQTWDQL LLHYQQEAKE IL SRGSTEA KITEVKVEPM TYLGSSQNEV LNTKPDYQKI LQNQSKVFDC MELVMDELQG SVKQLQAFMD ESTQCFQKVS VQL GKRSMQ QLDPSPARKL LKLQLQNPPA IHGSGSGSCQ HHHHHH UniProtKB: Kinetochore-associated protein DSN1 homolog |
-Macromolecule #6: Kinetochore scaffold 1
| Macromolecule | Name: Kinetochore scaffold 1 / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 38.644453 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GAMGGSMGKL VQSAQNEREK LQIKIDEMDK ILKKIDNCLT EMETETKNLE DEEKNNPVEE WDSEMRAAEK ELEQLKTEEE ELQRNLLEL EVQKEQTLAQ IDFMQKQRNR TEELLDQLSL SEWDVVEWSD DQAVFTFVYD TIQLTITFEE SVVGFPFLDK R YRKIVDVN ...String: GAMGGSMGKL VQSAQNEREK LQIKIDEMDK ILKKIDNCLT EMETETKNLE DEEKNNPVEE WDSEMRAAEK ELEQLKTEEE ELQRNLLEL EVQKEQTLAQ IDFMQKQRNR TEELLDQLSL SEWDVVEWSD DQAVFTFVYD TIQLTITFEE SVVGFPFLDK R YRKIVDVN FQSLLDEDQA PPSSLLVHKL IFQYVEEKES WKKTCTTQHQ LPKMLEEFSL VVHHCRLLGE EIEYLKRWGP NY NLMNIDI NNNELRLLFS SSAAFAKFEI TLFLSAYYPS VPLPSTIQNH VGNTSQDDIA TILSKVPLEN NYLKNVVKQI YQD LFQDCH FYH UniProtKB: Outer kinetochore KNL1 complex subunit KNL1 |
-Macromolecule #7: Kinetochore-associated protein NSL1 homolog
| Macromolecule | Name: Kinetochore-associated protein NSL1 homolog / type: protein_or_peptide / ID: 7 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 32.208951 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MAGSPELVVL DPPWDKELAA GTESQALVSA TPREDFRVRC TSKRAVTEML QLCGRFVQKL GDALPEEIRE PALRDAQWTF ESAVQENIS INGQAWQEAS DNCFMDSDIK VLEDQFDEII VDIATKRKQY PRKILECVIK TIKAKQEILK QYHPVVHPLD L KYDPDPAP ...String: MAGSPELVVL DPPWDKELAA GTESQALVSA TPREDFRVRC TSKRAVTEML QLCGRFVQKL GDALPEEIRE PALRDAQWTF ESAVQENIS INGQAWQEAS DNCFMDSDIK VLEDQFDEII VDIATKRKQY PRKILECVIK TIKAKQEILK QYHPVVHPLD L KYDPDPAP HMENLKCRGE TVAKEISEAM KSLPALIEQG EGFSQVLRMQ PVIHLQRIHQ EVFSSCHRKP DAKPENFITQ IE TTPTETA SRKTSDMVLK RKQTKDCPQR KWYPLRPKKI NLDT UniProtKB: Kinetochore-associated protein NSL1 homolog |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 63.9 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)