[English] 日本語
 Yorodumi
Yorodumi- EMDB-18172: NMNAT1 core-bound RANBP9-TWA1-WDR26 module of WDR26-CTLH E3 ligase -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | NMNAT1 core-bound RANBP9-TWA1-WDR26 module of WDR26-CTLH E3 ligase | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | E3 ubiquitin ligase / CTLH / GID / NMNAT1 / YPEL5 / NAD / NADH / LIGASE | ||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||
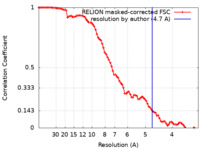
| Method | single particle reconstruction / cryo EM / Resolution: 4.7 Å | ||||||||||||
 Authors Authors | Chrustowicz J / Sherpa D / Schulman BA | ||||||||||||
| Funding support |  Germany, European Union, 3 items Germany, European Union, 3 items
| ||||||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2024 Journal: Mol Cell / Year: 2024Title: Non-canonical substrate recognition by the human WDR26-CTLH E3 ligase regulates prodrug metabolism. Authors: Karthik V Gottemukkala / Jakub Chrustowicz / Dawafuti Sherpa / Sara Sepic / Duc Tung Vu / Özge Karayel / Eleftheria C Papadopoulou / Annette Gross / Kenji Schorpp / Susanne von Gronau / ...Authors: Karthik V Gottemukkala / Jakub Chrustowicz / Dawafuti Sherpa / Sara Sepic / Duc Tung Vu / Özge Karayel / Eleftheria C Papadopoulou / Annette Gross / Kenji Schorpp / Susanne von Gronau / Kamyar Hadian / Peter J Murray / Matthias Mann / Brenda A Schulman / Arno F Alpi /  Abstract: The yeast glucose-induced degradation-deficient (GID) E3 ubiquitin ligase forms a suite of complexes with interchangeable receptors that selectively recruit N-terminal degron motifs of metabolic ...The yeast glucose-induced degradation-deficient (GID) E3 ubiquitin ligase forms a suite of complexes with interchangeable receptors that selectively recruit N-terminal degron motifs of metabolic enzyme substrates. The orthologous higher eukaryotic C-terminal to LisH (CTLH) E3 complex has been proposed to also recognize substrates through an alternative subunit, WDR26, which promotes the formation of supramolecular CTLH E3 assemblies. Here, we discover that human WDR26 binds the metabolic enzyme nicotinamide/nicotinic-acid-mononucleotide-adenylyltransferase 1 (NMNAT1) and mediates its CTLH E3-dependent ubiquitylation independently of canonical GID/CTLH E3-family substrate receptors. The CTLH subunit YPEL5 inhibits NMNAT1 ubiquitylation and cellular turnover by WDR26-CTLH E3, thereby affecting NMNAT1-mediated metabolic activation and cytotoxicity of the prodrug tiazofurin. Cryoelectron microscopy (cryo-EM) structures of NMNAT1- and YPEL5-bound WDR26-CTLH E3 complexes reveal an internal basic degron motif of NMNAT1 essential for targeting by WDR26-CTLH E3 and degron mimicry by YPEL5's N terminus antagonizing substrate binding. Thus, our data provide a mechanistic understanding of how YPEL5-WDR26-CTLH E3 acts as a modulator of NMNAT1-dependent metabolism. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18172.map.gz emd_18172.map.gz | 4.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18172-v30.xml emd-18172-v30.xml emd-18172.xml emd-18172.xml | 16.3 KB 16.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_18172_fsc.xml emd_18172_fsc.xml | 8.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_18172.png emd_18172.png | 114.4 KB | ||
| Masks |  emd_18172_msk_1.map emd_18172_msk_1.map | 52.7 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-18172.cif.gz emd-18172.cif.gz | 4.2 KB | ||
| Others |  emd_18172_additional_1.map.gz emd_18172_additional_1.map.gz emd_18172_half_map_1.map.gz emd_18172_half_map_1.map.gz emd_18172_half_map_2.map.gz emd_18172_half_map_2.map.gz | 47.1 MB 39.8 MB 39.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18172 http://ftp.pdbj.org/pub/emdb/structures/EMD-18172 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18172 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18172 | HTTPS FTP |
-Validation report
| Summary document |  emd_18172_validation.pdf.gz emd_18172_validation.pdf.gz | 826.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_18172_full_validation.pdf.gz emd_18172_full_validation.pdf.gz | 826 KB | Display | |
| Data in XML |  emd_18172_validation.xml.gz emd_18172_validation.xml.gz | 13.9 KB | Display | |
| Data in CIF |  emd_18172_validation.cif.gz emd_18172_validation.cif.gz | 19.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18172 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18172 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18172 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18172 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_18172.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18172.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.674 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_18172_msk_1.map emd_18172_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Map sharpened with DeepEMhancer
| File | emd_18172_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map sharpened with DeepEMhancer | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_18172_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_18172_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Complex of human WDR26-CTLH E3 ligase bound to NMNAT1
| Entire | Name: Complex of human WDR26-CTLH E3 ligase bound to NMNAT1 |
|---|---|
| Components |
|
-Supramolecule #1: Complex of human WDR26-CTLH E3 ligase bound to NMNAT1
| Supramolecule | Name: Complex of human WDR26-CTLH E3 ligase bound to NMNAT1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 Details: Map obtained by focused refinement over NMNAT1 core-bound RANBP9-TWA1-WDR26 module |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 1.1 MDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
| Details | Sample mixed with 0.01% beta-OG right before plunging |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 71.4 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.3000000000000003 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)