+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
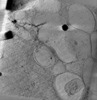
| Title | phagophore in fibrillar polyQ | |||||||||
 Map data Map data | phagphore with fibrillar polyQ | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | polyQ aggregates / neurodegeneration / PROTEIN FIBRIL | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | electron tomography / cryo EM | |||||||||
 Authors Authors | Zhao DY | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2024 Journal: Mol Cell / Year: 2024Title: Autophagy preferentially degrades non-fibrillar polyQ aggregates. Authors: Dorothy Y Zhao / Felix J B Bäuerlein / Itika Saha / F Ulrich Hartl / Wolfgang Baumeister / Florian Wilfling /   Abstract: Aggregation of proteins containing expanded polyglutamine (polyQ) repeats is the cytopathologic hallmark of a group of dominantly inherited neurodegenerative diseases, including Huntington's disease ...Aggregation of proteins containing expanded polyglutamine (polyQ) repeats is the cytopathologic hallmark of a group of dominantly inherited neurodegenerative diseases, including Huntington's disease (HD). Huntingtin (Htt), the disease protein of HD, forms amyloid-like fibrils by liquid-to-solid phase transition. Macroautophagy has been proposed to clear polyQ aggregates, but the efficiency of aggrephagy is limited. Here, we used cryo-electron tomography to visualize the interactions of autophagosomes with polyQ aggregates in cultured cells in situ. We found that an amorphous aggregate phase exists next to the radially organized polyQ fibrils. Autophagosomes preferentially engulfed this amorphous material, mediated by interactions between the autophagy receptor p62/SQSTM1 and the non-fibrillar aggregate surface. In contrast, amyloid fibrils excluded p62 and evaded clearance, resulting in trapping of autophagic structures. These results suggest that the limited efficiency of autophagy in clearing polyQ aggregates is due to the inability of autophagosomes to interact productively with the non-deformable, fibrillar disease aggregates. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18114.map.gz emd_18114.map.gz | 414.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18114-v30.xml emd-18114-v30.xml emd-18114.xml emd-18114.xml | 8.3 KB 8.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_18114.png emd_18114.png | 251.8 KB | ||
| Filedesc metadata |  emd-18114.cif.gz emd-18114.cif.gz | 3.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18114 http://ftp.pdbj.org/pub/emdb/structures/EMD-18114 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18114 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18114 | HTTPS FTP |
-Validation report
| Summary document |  emd_18114_validation.pdf.gz emd_18114_validation.pdf.gz | 404.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_18114_full_validation.pdf.gz emd_18114_full_validation.pdf.gz | 404.1 KB | Display | |
| Data in XML |  emd_18114_validation.xml.gz emd_18114_validation.xml.gz | 4.4 KB | Display | |
| Data in CIF |  emd_18114_validation.cif.gz emd_18114_validation.cif.gz | 4.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18114 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18114 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18114 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18114 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_18114.map.gz / Format: CCP4 / Size: 446.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18114.map.gz / Format: CCP4 / Size: 446.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | phagphore with fibrillar polyQ | ||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 12.76 Å | ||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : polyQ aggregates in Hek293 cells
| Entire | Name: polyQ aggregates in Hek293 cells |
|---|---|
| Components |
|
-Supramolecule #1: polyQ aggregates in Hek293 cells
| Supramolecule | Name: polyQ aggregates in Hek293 cells / type: cell / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | electron tomography |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 / Details: Cell media with 10% glycerol |
|---|---|
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 25 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK IV |
| Cryo protectant | 10% glycerol |
| Sectioning | Focused ion beam - Instrument: OTHER / Focused ion beam - Ion: OTHER / Focused ion beam - Voltage: 30 / Focused ion beam - Current: 0.05 / Focused ion beam - Duration: 1800 / Focused ion beam - Temperature: 100 K / Focused ion beam - Initial thickness: 1000 / Focused ion beam - Final thickness: 250 Focused ion beam - Details: The value given for _em_focused_ion_beam.instrument is FEI. This is not in a list of allowed values {'OTHER', 'DB235'} so OTHER is written into the XML file. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 2.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 5.0 µm / Nominal defocus min: 3.0 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Number images used: 50 |
|---|
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)
















