+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Wait Complex: Lateral open BAM bound Extended SurA | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Outer Membrane / Complex / Chaperone / Protein Folding / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology information: / maintenance of unfolded protein / Bam protein complex / Gram-negative-bacterium-type cell outer membrane assembly / Secretion of toxins / : / protein insertion into membrane / peptide binding / peptidylprolyl isomerase / peptidyl-prolyl cis-trans isomerase activity ...: / maintenance of unfolded protein / Bam protein complex / Gram-negative-bacterium-type cell outer membrane assembly / Secretion of toxins / : / protein insertion into membrane / peptide binding / peptidylprolyl isomerase / peptidyl-prolyl cis-trans isomerase activity / cell outer membrane / unfolded protein binding / protein folding / outer membrane-bounded periplasmic space / protein-macromolecule adaptor activity / cell adhesion / protein stabilization / response to antibiotic / cell surface / identical protein binding / membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
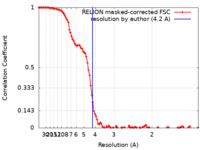
| Method | single particle reconstruction / cryo EM / Resolution: 4.2 Å | |||||||||
 Authors Authors | Fenn KL / Ranson NA | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Outer membrane protein assembly mediated by BAM-SurA complexes. Authors: Katherine L Fenn / Jim E Horne / Joel A Crossley / Nils Böhringer / Romany J Horne / Till F Schäberle / Antonio N Calabrese / Sheena E Radford / Neil A Ranson /   Abstract: The outer membrane is a formidable barrier that protects Gram-negative bacteria against environmental threats. Its integrity requires the correct folding and insertion of outer membrane proteins ...The outer membrane is a formidable barrier that protects Gram-negative bacteria against environmental threats. Its integrity requires the correct folding and insertion of outer membrane proteins (OMPs) by the membrane-embedded β-barrel assembly machinery (BAM). Unfolded OMPs are delivered to BAM by the periplasmic chaperone SurA, but how SurA and BAM work together to ensure successful OMP delivery and folding remains unclear. Here, guided by AlphaFold2 models, we use disulphide bond engineering in an attempt to trap SurA in the act of OMP delivery to BAM, and solve cryoEM structures of a series of complexes. The results suggest that SurA binds BAM at its soluble POTRA-1 domain, which may trigger conformational changes in both BAM and SurA that enable transfer of the unfolded OMP to the BAM lateral gate for insertion into the outer membrane. Mutations that disrupt the interaction between BAM and SurA result in outer membrane assembly defects, supporting the key role of SurA in outer membrane biogenesis. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18035.map.gz emd_18035.map.gz | 12.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18035-v30.xml emd-18035-v30.xml emd-18035.xml emd-18035.xml | 24.6 KB 24.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_18035_fsc.xml emd_18035_fsc.xml | 10.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_18035.png emd_18035.png | 147.4 KB | ||
| Filedesc metadata |  emd-18035.cif.gz emd-18035.cif.gz | 7.6 KB | ||
| Others |  emd_18035_half_map_1.map.gz emd_18035_half_map_1.map.gz emd_18035_half_map_2.map.gz emd_18035_half_map_2.map.gz | 80.7 MB 80.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18035 http://ftp.pdbj.org/pub/emdb/structures/EMD-18035 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18035 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18035 | HTTPS FTP |
-Related structure data
| Related structure data |  8pz2MC  8pz1C  8pzuC  8pzvC  8q0gC  8qp5C  8qpuC  8qpvC  8qpwC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_18035.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18035.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.71 Å | ||||||||||||||||||||||||||||||||||||
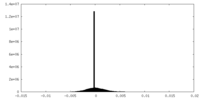
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_18035_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
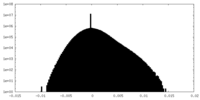
| Density Histograms |
-Half map: #2
| File | emd_18035_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Wait Complex: Lateral open BAM bound Extended SurA
| Entire | Name: Wait Complex: Lateral open BAM bound Extended SurA |
|---|---|
| Components |
|
-Supramolecule #1: Wait Complex: Lateral open BAM bound Extended SurA
| Supramolecule | Name: Wait Complex: Lateral open BAM bound Extended SurA / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: BamA POTRA1 disulphide bonded to SurA Core domain (BamA R76C, SurA K27C) |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 250 KDa |
-Macromolecule #1: Outer membrane protein assembly factor BamA
| Macromolecule | Name: Outer membrane protein assembly factor BamA / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 88.460688 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: AEGFVVKDIH FEGLQRVAVG AALLSMPVRT GDTVNDEDIS NTIRALFATG NFEDVCVLRD GDTLLVQVKE RPTIASITFS GNKSVKDDM LKQNLEASGV RVGESLDRTT IADIEKGLED FYYSVGKYSA SVKAVVTPLP RNRVDLKLVF QEGVSAEIQQ I NIVGNHAF ...String: AEGFVVKDIH FEGLQRVAVG AALLSMPVRT GDTVNDEDIS NTIRALFATG NFEDVCVLRD GDTLLVQVKE RPTIASITFS GNKSVKDDM LKQNLEASGV RVGESLDRTT IADIEKGLED FYYSVGKYSA SVKAVVTPLP RNRVDLKLVF QEGVSAEIQQ I NIVGNHAF TTDELISHFQ LRDEVPWWNV VGDRKYQKQK LAGDLETLRS YYLDRGYARF NIDSTQVSLT PDKKGIYVTV NI TEGDQYK LSGVEVSGNL AGHSAEIEQL TKIEPGELYN GTKVTKMEDD IKKLLGRYGY AYPRVQSMPE INDADKTVKL RVN VDAGNR FYVRKIRFEG NDTSKDAVLR REMRQMEGAW LGSDLVDQGK ERLNRLGFFE TVDTDTQRVP GSPDQVDVVY KVKE RNTGS FNFGIGYGTE SGVSFQAGVQ QDNWLGTGYA VGINGTKNDY QTYAELSVTN PYFTVDGVSL GGRLFYNDFQ ADDAD LSDY TNKSYGTDVT LGFPINEYNS LRAGLGYVHN SLSNMQPQVA MWRYLYSMGE HPSTSDQDNS FKTDDFTFNY GWTYNK LDR GYFPTDGSRV NLTGKVTIPG SDNEYYKVTL DTATYVPIDD DHKWVVLGRT RWGYGDGLGG KEMPFYENFY AGGSSTV RG FQSNTIGPKA VYFPHQASNY DPDYDYECAT QDGAKDLCKS DDAVGGNAMA VASLEFITPT PFISDKYANS VRTSFFWD M GTVWDTNWDS SQYSGYPDYS DPSNIRMSAG IALQWMSPLG PLVFSYAQPF KKYDGDKAEQ FQFNIGKTW UniProtKB: Outer membrane protein assembly factor BamA |
-Macromolecule #2: Outer membrane protein assembly factor BamB
| Macromolecule | Name: Outer membrane protein assembly factor BamB / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 39.882375 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: CSLFNSEEDV VKMSPLPTVE NQFTPTTAWS TSVGSGIGNF YSNLHPALAD NVVYAADRAG LVKALNADDG KEIWSVSLAE KDGWFSKEP ALLSGGVTVS GGHVYIGSEK AQVYALNTSD GTVAWQTKVA GEALSRPVVS DGLVLIHTSN GQLQALNEAD G AVKWTVNL ...String: CSLFNSEEDV VKMSPLPTVE NQFTPTTAWS TSVGSGIGNF YSNLHPALAD NVVYAADRAG LVKALNADDG KEIWSVSLAE KDGWFSKEP ALLSGGVTVS GGHVYIGSEK AQVYALNTSD GTVAWQTKVA GEALSRPVVS DGLVLIHTSN GQLQALNEAD G AVKWTVNL DMPSLSLRGE SAPTTAFGAA VVGGDNGRVS AVLMEQGQMI WQQRISQATG STEIDRLSDV DTTPVVVNGV VF ALAYNGN LTALDLRSGQ IMWKRELGSV NDFIVDGNRI YLVDQNDRVM ALTIDGGVTL WTQSDLLHRL LTSPVLYNGN LVV GDSEGY LHWINVEDGR FVAQQKVDSS GFQTEPVAAD GKLLIQAKDG TVYSITR UniProtKB: Outer membrane protein assembly factor BamB |
-Macromolecule #3: Outer membrane protein assembly factor BamC
| Macromolecule | Name: Outer membrane protein assembly factor BamC / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 34.40125 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: CSSDSRYKRQ VSGDEAYLEA APLAELHAPA GMILPVTSGD YAIPVTNGSG AVGKALDIRP PAQPLALVSG ARTQFTGDTA SLLVENGRG NTLWPQVVSV LQAKNYTITQ RDDAGQTLTT DWVQWNRLDE DEQYRGRYQI SVKPQGYQQA VTVKLLNLEQ A GKPVADAA ...String: CSSDSRYKRQ VSGDEAYLEA APLAELHAPA GMILPVTSGD YAIPVTNGSG AVGKALDIRP PAQPLALVSG ARTQFTGDTA SLLVENGRG NTLWPQVVSV LQAKNYTITQ RDDAGQTLTT DWVQWNRLDE DEQYRGRYQI SVKPQGYQQA VTVKLLNLEQ A GKPVADAA SMQRYSTEMM NVISAGLDKS ATDAANAAQN RASTTMDVQS AADDTGLPML VVRGPFNVVW QRLPAALEKV GM KVTDSTR SQGNMAVTYK PLSDSDWQEL GASDPGLASG DYKLQVGDLD NRSSLQFIDP KGHTLTQSQN DALVAVFQAA FSK UniProtKB: Outer membrane protein assembly factor BamC |
-Macromolecule #4: Outer membrane protein assembly factor BamD
| Macromolecule | Name: Outer membrane protein assembly factor BamD / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 25.816818 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: CSGSKEEVPD NPPNEIYATA QQKLQDGNWR QAITQLEALD NRYPFGPYSQ QVQLDLIYAY YKNADLPLAQ AAIDRFIRLN PTHPNIDYV MYMRGLTNMA LDDSALQGFF GVDRSDRDPQ HARAAFSDFS KLVRGYPNSQ YTTDATKRLV FLKDRLAKYE Y SVAEYYTE ...String: CSGSKEEVPD NPPNEIYATA QQKLQDGNWR QAITQLEALD NRYPFGPYSQ QVQLDLIYAY YKNADLPLAQ AAIDRFIRLN PTHPNIDYV MYMRGLTNMA LDDSALQGFF GVDRSDRDPQ HARAAFSDFS KLVRGYPNSQ YTTDATKRLV FLKDRLAKYE Y SVAEYYTE RGAWVAVVNR VEGMLRDYPD TQATRDALPL MENAYRQMQM NAQAEKVAKI IAANSSNT UniProtKB: Outer membrane protein assembly factor BamD |
-Macromolecule #5: Outer membrane protein assembly factor BamE
| Macromolecule | Name: Outer membrane protein assembly factor BamE / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 11.610833 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: CSTLERVVYR PDINQGNYLT ANDVSKIRVG MTQQQVAYAL GTPLMSDPFG TNTWFYVFRQ QPGHEGVTQQ TLTLTFNSSG VLTNIDNKP ALSGNGGHHH HHHHH UniProtKB: Outer membrane protein assembly factor BamE |
-Macromolecule #6: Chaperone SurA
| Macromolecule | Name: Chaperone SurA / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO / EC number: peptidylprolyl isomerase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 49.36427 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GSSAWSHPQF EKGGGSGGGS GGSAWSHPQF EKSSGENLYF QGAPQVVDCV AAVVNNGVVL ESDVDGLMQS VKLNAAQARQ QLPDDATLR HQIMERLIMD QIILQMGQKM GVKISDEQLD QAIANIAKQN NMTLDQMRSR LAYDGLNYNT YRNQIRKEMI I SEVRNNEV ...String: GSSAWSHPQF EKGGGSGGGS GGSAWSHPQF EKSSGENLYF QGAPQVVDCV AAVVNNGVVL ESDVDGLMQS VKLNAAQARQ QLPDDATLR HQIMERLIMD QIILQMGQKM GVKISDEQLD QAIANIAKQN NMTLDQMRSR LAYDGLNYNT YRNQIRKEMI I SEVRNNEV RRRITILPQE VESLAQQVGN QNDASTELNL SHILIPLPEN PTSDQVNEAE SQARAIVDQA RNGADFGKLA IA HSADQQA LNGGQMGWGR IQELPGIFAQ ALSTAKKGDI VGPIRSGVGF HILKVNDLRG ESKNISVTEV HARHILLKPS PIM TDEQAR VKLEQIAADI KSGKTTFAAA AKEFSQDPGS ANQGGDLGWA TPDIFDPAFR DALTRLNKGQ MSAPVHSSFG WHLI ELLDT RNVDKTDAAQ KDRAYRMLMN RKFSEEAASW MQEQRASAYV KILSN UniProtKB: Chaperone SurA |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 60 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: TFS Selectris / Energy filter - Slit width: 10 eV |
| Software | Name: EPU |
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Number grids imaged: 1 / Number real images: 15992 / Average electron dose: 37.9 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.9 µm / Nominal magnification: 165000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||
|---|---|---|---|---|---|---|---|
| Software | Name: UCSF ChimeraX / Details: Rigid Body Fit | ||||||
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Overall B value: 156 / Target criteria: Cross-correlation coefficient | ||||||
| Output model |  PDB-8pz2: |
 Movie
Movie Controller
Controller



















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)