+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of K27A mutant E.coli DPS | |||||||||
 Map data Map data | Structure of K27A mutant E.coli DPS determined by movement-free cryoEM. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ferritin / DNA / METAL BINDING PROTEIN | |||||||||
| Function / homology | :  Function and homology information Function and homology information | |||||||||
| Biological species |  | |||||||||
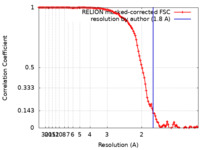
| Method | single particle reconstruction / cryo EM / Resolution: 1.8 Å | |||||||||
 Authors Authors | Dickerson JL / Russo CJ | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Ultramicroscopy / Year: 2024 Journal: Ultramicroscopy / Year: 2024Title: Accurate magnification determination for cryoEM using gold. Authors: Joshua L Dickerson / Erin Leahy / Mathew J Peet / Katerina Naydenova / Christopher J Russo /  Abstract: Determining the correct magnified pixel size of single-particle cryoEM micrographs is necessary to maximize resolution and enable accurate model building. Here we describe a simple and rapid ...Determining the correct magnified pixel size of single-particle cryoEM micrographs is necessary to maximize resolution and enable accurate model building. Here we describe a simple and rapid procedure for determining the absolute magnification in an electron cryomicroscope to a precision of <0.5%. We show how to use the atomic lattice spacings of crystals of thin and readily available test specimens, such as gold, as an absolute reference to determine magnification for both room temperature and cryogenic imaging. We compare this method to other commonly used methods, and show that it provides comparable accuracy in spite of its simplicity. This magnification calibration method provides a definitive reference quantity for data analysis and processing, simplifies the combination of multiple datasets from different microscopes and detectors, and improves the accuracy with which the contrast transfer function of the microscope can be determined. We also provide an open source program, magCalEM, which can be used to accurately estimate the magnified pixel size of a cryoEM dataset ex post facto. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17992.map.gz emd_17992.map.gz | 15.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17992-v30.xml emd-17992-v30.xml emd-17992.xml emd-17992.xml | 14.2 KB 14.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_17992_fsc.xml emd_17992_fsc.xml | 11.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_17992.png emd_17992.png | 271.8 KB | ||
| Masks |  emd_17992_msk_1.map emd_17992_msk_1.map | 125 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-17992.cif.gz emd-17992.cif.gz | 4.7 KB | ||
| Others |  emd_17992_half_map_1.map.gz emd_17992_half_map_1.map.gz emd_17992_half_map_2.map.gz emd_17992_half_map_2.map.gz | 97.8 MB 97.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17992 http://ftp.pdbj.org/pub/emdb/structures/EMD-17992 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17992 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17992 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_17992.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17992.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Structure of K27A mutant E.coli DPS determined by movement-free cryoEM. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.646 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_17992_msk_1.map emd_17992_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Unfiltered half map 1
| File | emd_17992_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unfiltered half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Unfiltered half map 2
| File | emd_17992_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unfiltered half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : DNA protection during starvation protein (DPS)
| Entire | Name: DNA protection during starvation protein (DPS) |
|---|---|
| Components |
|
-Supramolecule #1: DNA protection during starvation protein (DPS)
| Supramolecule | Name: DNA protection during starvation protein (DPS) / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 224 KDa |
-Macromolecule #1: DNA protection during starvation protein (DPS)
| Macromolecule | Name: DNA protection during starvation protein (DPS) / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Sequence | String: MSTAKLVKSK ATNLLYTRND VSDSEKAATV ELLNRQVIQF IDLSLITKQA HWNMRGANFI AVHEMLDGFR TALIDHLDTM AERAVQLGG VALGTTQVIN SKTPLKSYPL DIHNVQDHLK ELADRYAIVA NDVRKAIGEA KDDDTADILT AASRDLDKFL W FIESNIE UniProtKB: UNIPROTKB: POABT2 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.7 |
|---|---|
| Grid | Material: GOLD / Support film - Material: GOLD / Support film - topology: HOLEY ARRAY / Support film - Film thickness: 30 / Pretreatment - Type: PLASMA CLEANING |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.6 µm / Nominal defocus min: 0.6 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)