+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the Lifeact13-F-actin complex | |||||||||
 Map data Map data | Local resolution filtered map. Used for model building | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Cytoskeleton / Actin / Lifeact / labelling / STRUCTURAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationcytoskeletal motor activator activity / myosin heavy chain binding / tropomyosin binding / actin filament bundle / troponin I binding / filamentous actin / mesenchyme migration / skeletal muscle myofibril / actin filament bundle assembly / striated muscle thin filament ...cytoskeletal motor activator activity / myosin heavy chain binding / tropomyosin binding / actin filament bundle / troponin I binding / filamentous actin / mesenchyme migration / skeletal muscle myofibril / actin filament bundle assembly / striated muscle thin filament / skeletal muscle thin filament assembly / actin monomer binding / skeletal muscle fiber development / stress fiber / titin binding / actin filament polymerization / actin filament / filopodium / Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement / calcium-dependent protein binding / lamellipodium / cell body / protein domain specific binding / hydrolase activity / calcium ion binding / positive regulation of gene expression / magnesium ion binding / ATP binding / identical protein binding / cytoplasm Similarity search - Function | |||||||||
| Biological species |   | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.55 Å | |||||||||
 Authors Authors | Belyy A / Raunser S | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: To be published Authors: Belyy A / Raunser S | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17979.map.gz emd_17979.map.gz | 60.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17979-v30.xml emd-17979-v30.xml emd-17979.xml emd-17979.xml | 16.3 KB 16.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_17979.png emd_17979.png | 111.1 KB | ||
| Masks |  emd_17979_msk_1.map emd_17979_msk_1.map | 103 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-17979.cif.gz emd-17979.cif.gz | 5.8 KB | ||
| Others |  emd_17979_half_map_1.map.gz emd_17979_half_map_1.map.gz emd_17979_half_map_2.map.gz emd_17979_half_map_2.map.gz | 79.4 MB 79.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17979 http://ftp.pdbj.org/pub/emdb/structures/EMD-17979 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17979 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17979 | HTTPS FTP |
-Related structure data
| Related structure data |  8pvxM M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_17979.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17979.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Local resolution filtered map. Used for model building | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.21 Å | ||||||||||||||||||||||||||||||||||||
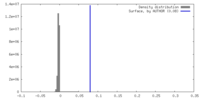
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_17979_msk_1.map emd_17979_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
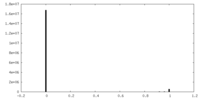
| Density Histograms |
-Half map: Second half map
| File | emd_17979_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Second half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
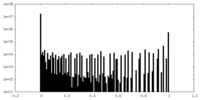
| Density Histograms |
-Half map: First half map
| File | emd_17979_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | First half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : The complex between skeletal actin from rabbit and Lifeact13
| Entire | Name: The complex between skeletal actin from rabbit and Lifeact13 |
|---|---|
| Components |
|
-Supramolecule #1: The complex between skeletal actin from rabbit and Lifeact13
| Supramolecule | Name: The complex between skeletal actin from rabbit and Lifeact13 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|
-Supramolecule #2: Lifeact13
| Supramolecule | Name: Lifeact13 / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #3: Actin, alpha skeletal muscle
| Supramolecule | Name: Actin, alpha skeletal muscle / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Lifeact13
| Macromolecule | Name: Lifeact13 / type: protein_or_peptide / ID: 1 / Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 1.452758 KDa |
| Sequence | String: MGVADLIKKF ESI |
-Macromolecule #2: Actin, alpha skeletal muscle
| Macromolecule | Name: Actin, alpha skeletal muscle / type: protein_or_peptide / ID: 2 / Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 42.109973 KDa |
| Sequence | String: MCDEDETTAL VCDNGSGLVK AGFAGDDAPR AVFPSIVGRP RHQGVMVGMG QKDSYVGDEA QSKRGILTLK YPIE(HIC)G IIT NWDDMEKIWH HTFYNELRVA PEEHPTLLTE APLNPKANRE KMTQIMFETF NVPAMYVAIQ AVLSLYASGR TTGIVLD SG DGVTHNVPIY ...String: MCDEDETTAL VCDNGSGLVK AGFAGDDAPR AVFPSIVGRP RHQGVMVGMG QKDSYVGDEA QSKRGILTLK YPIE(HIC)G IIT NWDDMEKIWH HTFYNELRVA PEEHPTLLTE APLNPKANRE KMTQIMFETF NVPAMYVAIQ AVLSLYASGR TTGIVLD SG DGVTHNVPIY EGYALPHAIM RLDLAGRDLT DYLMKILTER GYSFVTTAER EIVRDIKEKL CYVALDFENE MATAASSS S LEKSYELPDG QVITIGNERF RCPETLFQPS FIGMESAGIH ETTYNSIMKC DIDIRKDLYA NNVMSGGTTM YPGIADRMQ KEITALAPST MKIKIIAPPE RKYSVWIGGS ILASLSTFQQ MWITKQEYDE AGPSIVHRKC F UniProtKB: Actin, alpha skeletal muscle |
-Macromolecule #3: ADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 3 / Number of copies: 5 / Formula: ADP |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-Macromolecule #4: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 4 / Number of copies: 5 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: INTEGRATING / Number grids imaged: 1 / Average exposure time: 3.0 sec. / Average electron dose: 56.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: EMDB MAP EMDB ID: |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 3.55 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 1371447 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Output model |  PDB-8pvx: |
-Atomic model buiding 2
| Refinement | Protocol: FLEXIBLE FIT |
|---|---|
| Output model |  PDB-8pvx: |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)