[English] 日本語
 Yorodumi
Yorodumi- EMDB-17928: Structure of BARD1 ARD-BRCTs in complex with H2AKc15ub nucleosome... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
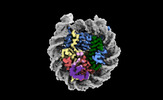
| Title | Structure of BARD1 ARD-BRCTs in complex with H2AKc15ub nucleosomes (Map 2) | |||||||||||||||
 Map data Map data | ||||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | BRCA1-BARD1 / chromatin recognition / nucleosomes / ubiquitin / DNA BINDING PROTEIN | |||||||||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.75 Å | |||||||||||||||
 Authors Authors | Foglizzo M / Burdett H / Wilson MD / Zeqiraj E | |||||||||||||||
| Funding support |  United Kingdom, 4 items United Kingdom, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nucleic Acids Res / Year: 2023 Journal: Nucleic Acids Res / Year: 2023Title: BRCA1-BARD1 combines multiple chromatin recognition modules to bridge nascent nucleosomes. Authors: Hayden Burdett / Martina Foglizzo / Laura J Musgrove / Dhananjay Kumar / Gillian Clifford / Lisa J Campbell / George R Heath / Elton Zeqiraj / Marcus D Wilson /  Abstract: Chromatin association of the BRCA1-BARD1 heterodimer is critical to promote homologous recombination repair of DNA double-strand breaks (DSBs) in S/G2. How the BRCA1-BARD1 complex interacts with ...Chromatin association of the BRCA1-BARD1 heterodimer is critical to promote homologous recombination repair of DNA double-strand breaks (DSBs) in S/G2. How the BRCA1-BARD1 complex interacts with chromatin that contains both damage induced histone H2A ubiquitin and inhibitory H4K20 methylation is not fully understood. We characterised BRCA1-BARD1 binding and enzymatic activity to an array of mono- and di-nucleosome substrates using biochemical, structural and single molecule imaging approaches. We found that the BRCA1-BARD1 complex preferentially interacts and modifies di-nucleosomes over mono-nucleosomes, allowing integration of H2A Lys-15 ubiquitylation signals with other chromatin modifications and features. Using high speed- atomic force microscopy (HS-AFM) to monitor how the BRCA1-BARD1 complex recognises chromatin in real time, we saw a highly dynamic complex that bridges two nucleosomes and associates with the DNA linker region. Bridging is aided by multivalent cross-nucleosome interactions that enhance BRCA1-BARD1 E3 ubiquitin ligase catalytic activity. Multivalent interactions across nucleosomes explain how BRCA1-BARD1 can recognise chromatin that retains partial di-methylation at H4 Lys-20 (H4K20me2), a parental histone mark that blocks BRCA1-BARD1 interaction with nucleosomes, to promote its enzymatic and DNA repair activities. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17928.map.gz emd_17928.map.gz | 117.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17928-v30.xml emd-17928-v30.xml emd-17928.xml emd-17928.xml | 23.8 KB 23.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_17928.png emd_17928.png | 49.2 KB | ||
| Masks |  emd_17928_msk_1.map emd_17928_msk_1.map | 125 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-17928.cif.gz emd-17928.cif.gz | 5.1 KB | ||
| Others |  emd_17928_additional_1.map.gz emd_17928_additional_1.map.gz emd_17928_additional_2.map.gz emd_17928_additional_2.map.gz emd_17928_half_map_1.map.gz emd_17928_half_map_1.map.gz emd_17928_half_map_2.map.gz emd_17928_half_map_2.map.gz | 62 MB 111.6 MB 116 MB 116 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17928 http://ftp.pdbj.org/pub/emdb/structures/EMD-17928 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17928 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17928 | HTTPS FTP |
-Validation report
| Summary document |  emd_17928_validation.pdf.gz emd_17928_validation.pdf.gz | 850 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_17928_full_validation.pdf.gz emd_17928_full_validation.pdf.gz | 849.6 KB | Display | |
| Data in XML |  emd_17928_validation.xml.gz emd_17928_validation.xml.gz | 13.9 KB | Display | |
| Data in CIF |  emd_17928_validation.cif.gz emd_17928_validation.cif.gz | 16.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17928 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17928 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17928 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17928 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_17928.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17928.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.86 Å | ||||||||||||||||||||||||||||||||||||
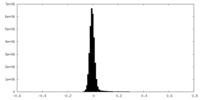
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_17928_msk_1.map emd_17928_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
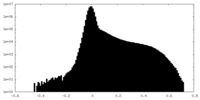
| Density Histograms |
-Additional map: #1
| File | emd_17928_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: #2
| File | emd_17928_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_17928_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_17928_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : BARD1 ARD-BRCTs in complex with H2AKc15ub nucleosomes
| Entire | Name: BARD1 ARD-BRCTs in complex with H2AKc15ub nucleosomes |
|---|---|
| Components |
|
-Supramolecule #1: BARD1 ARD-BRCTs in complex with H2AKc15ub nucleosomes
| Supramolecule | Name: BARD1 ARD-BRCTs in complex with H2AKc15ub nucleosomes / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#7 |
|---|---|
| Molecular weight | Theoretical: 246 KDa |
-Supramolecule #2: DNA, Histones and Polyubiquitin-B
| Supramolecule | Name: DNA, Histones and Polyubiquitin-B / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#5, #7 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #3: BRCA1 associated RING domain 1
| Supramolecule | Name: BRCA1 associated RING domain 1 / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #6 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
Details: 20 mM HEPES pH 7.5, 50 mM NaCl and 1 mM DTT | |||||||||
| Grid | Model: Quantifoil R3.5/1 / Material: COPPER / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR Details: Quantifoil R3.5/1 200-mesh grids (Quantifoil Micro Tools GmbH) were glow-discharged for 30 s at 40 mA using a GloQube (Quorum) glow discharge unit | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV / Details: blot force = 0 N blot time = 8 s. | |||||||||
| Details | Purified cat BRCA1dExon11-FL BARD1 (at 3 uM) was incubated with H2AKc15ub mono-nucleosomes (at 1.5 uM) for 1 h on ice before cryo-EM grids preparation |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Number grids imaged: 1 / Number real images: 16015 / Average exposure time: 5.0 sec. / Average electron dose: 36.37 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.1 µm / Nominal defocus min: 1.7 µm / Nominal magnification: 96000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)