+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Chaetomium thermophilum Rix1-complex | |||||||||
 Map data Map data | Chaetomium thermophilum Rix1-complex - local resolution filtered | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | biogenesis / pre-60S / 5S RNP / RIBOSOME | |||||||||
| Function / homology |  Function and homology information Function and homology informationrixosome complex / nuclear pre-replicative complex / DNA-templated DNA replication / rRNA processing / nucleus Similarity search - Function | |||||||||
| Biological species |  Thermochaetoides thermophila DSM 1495 (fungus) Thermochaetoides thermophila DSM 1495 (fungus) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.91 Å | |||||||||
 Authors Authors | Thoms M / Cheng J / Denk T / Berninghausen O / Beckmann R | |||||||||
| Funding support | European Union, 1 items
| |||||||||
 Citation Citation |  Journal: EMBO Rep / Year: 2023 Journal: EMBO Rep / Year: 2023Title: Structural insights into coordinating 5S RNP rotation with ITS2 pre-RNA processing during ribosome formation. Authors: Matthias Thoms / Benjamin Lau / Jingdong Cheng / Lisa Fromm / Timo Denk / Nikola Kellner / Dirk Flemming / Paulina Fischer / Laurent Falquet / Otto Berninghausen / Roland Beckmann / Ed Hurt /    Abstract: The rixosome defined in Schizosaccharomyces pombe and humans performs diverse roles in pre-ribosomal RNA processing and gene silencing. Here, we isolate and describe the conserved rixosome from ...The rixosome defined in Schizosaccharomyces pombe and humans performs diverse roles in pre-ribosomal RNA processing and gene silencing. Here, we isolate and describe the conserved rixosome from Chaetomium thermophilum, which consists of two sub-modules, the sphere-like Rix1-Ipi3-Ipi1 and the butterfly-like Las1-Grc3 complex, connected by a flexible linker. The Rix1 complex of the rixosome utilizes Sda1 as landing platform on nucleoplasmic pre-60S particles to wedge between the 5S rRNA tip and L1-stalk, thereby facilitating the 180° rotation of the immature 5S RNP towards its mature conformation. Upon rixosome positioning, the other sub-module with Las1 endonuclease and Grc3 polynucleotide-kinase can reach a strategic position at the pre-60S foot to cleave and 5' phosphorylate the nearby ITS2 pre-rRNA. Finally, inward movement of the L1 stalk permits the flexible Nop53 N-terminus with its AIM motif to become positioned at the base of the L1-stalk to facilitate Mtr4 helicase-exosome participation for completing ITS2 removal. Thus, the rixosome structure elucidates the coordination of two central ribosome biogenesis events, but its role in gene silencing may adapt similar strategies. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17879.map.gz emd_17879.map.gz | 2.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17879-v30.xml emd-17879-v30.xml emd-17879.xml emd-17879.xml | 26.1 KB 26.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_17879.png emd_17879.png | 119.6 KB | ||
| Filedesc metadata |  emd-17879.cif.gz emd-17879.cif.gz | 6.1 KB | ||
| Others |  emd_17879_additional_1.map.gz emd_17879_additional_1.map.gz emd_17879_additional_2.map.gz emd_17879_additional_2.map.gz emd_17879_additional_3.map.gz emd_17879_additional_3.map.gz emd_17879_additional_4.map.gz emd_17879_additional_4.map.gz emd_17879_additional_5.map.gz emd_17879_additional_5.map.gz emd_17879_half_map_1.map.gz emd_17879_half_map_1.map.gz emd_17879_half_map_2.map.gz emd_17879_half_map_2.map.gz | 41.5 MB 2.6 MB 41.8 MB 77.8 MB 77.8 MB 77.6 MB 77.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17879 http://ftp.pdbj.org/pub/emdb/structures/EMD-17879 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17879 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17879 | HTTPS FTP |
-Related structure data
| Related structure data |  8ptwMC  8puwC  8pv1C  8pv2C  8pv3C  8pv4C  8pv5C  8pv6C  8pv7C  8pv8C  8pvkC  8pvlC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_17879.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17879.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Chaetomium thermophilum Rix1-complex - local resolution filtered | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.045 Å | ||||||||||||||||||||||||||||||||||||
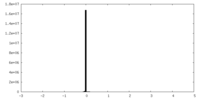
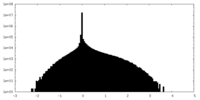
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Chaetomium thermophilum Rix1-complex
| File | emd_17879_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Chaetomium thermophilum Rix1-complex | ||||||||||||
| Projections & Slices |
| ||||||||||||
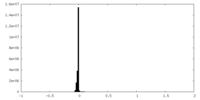
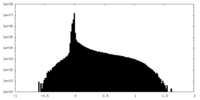
| Density Histograms |
-Additional map: Chaetomium thermophilum Rix1-complex - C1 symmetry - local...
| File | emd_17879_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Chaetomium thermophilum Rix1-complex - C1 symmetry - local resolution filtered | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Chaetomium thermophilum Rix1-complex - C1 symmetry
| File | emd_17879_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Chaetomium thermophilum Rix1-complex - C1 symmetry | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Chaetomium thermophilum Rix1-complex - C1 symmetry - half map A
| File | emd_17879_additional_4.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Chaetomium thermophilum Rix1-complex - C1 symmetry - half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Chaetomium thermophilum Rix1-complex - C1 symmetry - half map B
| File | emd_17879_additional_5.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Chaetomium thermophilum Rix1-complex - C1 symmetry - half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Chaetomium thermophilum Rix1-complex - half map A
| File | emd_17879_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Chaetomium thermophilum Rix1-complex - half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Chaetomium thermophilum Rix1-complex - half map B
| File | emd_17879_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Chaetomium thermophilum Rix1-complex - half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Chaetomium thermophilum Rix1-complex
| Entire | Name: Chaetomium thermophilum Rix1-complex |
|---|---|
| Components |
|
-Supramolecule #1: Chaetomium thermophilum Rix1-complex
| Supramolecule | Name: Chaetomium thermophilum Rix1-complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Thermochaetoides thermophila DSM 1495 (fungus) Thermochaetoides thermophila DSM 1495 (fungus) |
-Macromolecule #1: Pre-rRNA-processing protein IPI3
| Macromolecule | Name: Pre-rRNA-processing protein IPI3 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Thermochaetoides thermophila DSM 1495 (fungus) Thermochaetoides thermophila DSM 1495 (fungus) |
| Molecular weight | Theoretical: 47.627816 KDa |
| Sequence | String: MLTEEFVSAI CGPPLSSNTA IAKDVGIYCH TLSPSYSVKS TFKKSSVPVN CLAVSDTHIF AGQHEKAYVH VYSRLRGNQE AFVALPERI RCLILIGDIL VVGTTEGRLM LWEICTGRLV STPARHVQAV SCVAATPSHV LTGSDDSDIH VWSLSQLLEL D SAAEHEPL ...String: MLTEEFVSAI CGPPLSSNTA IAKDVGIYCH TLSPSYSVKS TFKKSSVPVN CLAVSDTHIF AGQHEKAYVH VYSRLRGNQE AFVALPERI RCLILIGDIL VVGTTEGRLM LWEICTGRLV STPARHVQAV SCVAATPSHV LTGSDDSDIH VWSLSQLLEL D SAAEHEPL RTLANHRAAI TALAVSPSDS ADTNFCVSAS KDKSCIIWNY QTGDALRTLI FPGYPLCMSL DPSSRAIFVS CE DSSLYVA EMFGEKPLLG PGSEDPSTVV QISTPFGATQ PDVGPASCLS VSYDGTMLLT GHPRGQIMRW DISENKSPVE LAN LNAAVT NLIFVSPFLT SKPTKTVNII KPSQAERAYT FTAQFEPMSF TKSRLDSLLN ATGFPADALE SAIVAFYQPV TQSA GDQEL QRQNEELWEI INEQRALQKE TLQRYVEAKS SR UniProtKB: Pre-rRNA-processing protein IPI3 |
-Macromolecule #2: Pre-rRNA-processing protein RIX1
| Macromolecule | Name: Pre-rRNA-processing protein RIX1 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Thermochaetoides thermophila DSM 1495 (fungus) Thermochaetoides thermophila DSM 1495 (fungus) |
| Molecular weight | Theoretical: 84.849219 KDa |
| Sequence | String: MTAPPDLRVV CHRLASTPVD SLPRLCPLLI NHVLRCGGPL SEPQDAKGKD RTSETAMLVH KFRTHITSLL TGKSPAGRFT AVCLIKAVI DVGGWESLRS AEPWIRGLIG VLQKPDPLSS KELSIVTLTK LYILLQDYQT LIREMATPTL PGYATACLQL I KPPASGRP ...String: MTAPPDLRVV CHRLASTPVD SLPRLCPLLI NHVLRCGGPL SEPQDAKGKD RTSETAMLVH KFRTHITSLL TGKSPAGRFT AVCLIKAVI DVGGWESLRS AEPWIRGLIG VLQKPDPLSS KELSIVTLTK LYILLQDYQT LIREMATPTL PGYATACLQL I KPPASGRP LKVPLNFVDT VAWSLSKLVV LYSTTMRPFS GQIKSALRPY IAPTSSDNVV VPQSLKENSR NLLILLTYTA PK NGSSDEW VKAIRATILD CHTTADQVFR AVRESWESTT GYHIQPVNAT GEPSGGGDSV DELPPWSGLQ AGAERLTGLL EYL TAYFNN PTRAPVNVPL GELLDLTTRL TLVIPPSLGA EDSIETNPAI GRDEKAELWS ALPDIHHAVL RLHCAIIRRL EANA IPLAT DIIDQMVRVS TASKQLPSVR ETAYILAKEI LLLAGSTLPK LTVDILIPLI QSSCHDILTA AGHAQPAQSQ SSVPV TASK QQKSSSPALT NADAFLPGQS SSSTPKTSTA SPVSQAASAL LPTFFTHLPQ KHLPPDIRGL LDRTAILSHN QSAMLA SCL HPYRDSRGRY YPSILPFLVR RFPRDESVEV LRSNLVKVGG SDASRGWDLS NGVTRDISYG REFAQEMISE EKGVVKE DE TFAKEIEPVK STAKPATSAN AWGVEMELDV EHVNVAPIPE TTNPFATVVG TTSQPSTLIQ PACPSSPLKR KSDAEEFD E GSRPKRVDTG KAVSHPQMAV ISSVPKPEED KSDESSDSEG SVQIDMTLED DEEDEEEEDE UniProtKB: Pre-rRNA-processing protein RIX1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 43.6 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.5 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.91 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 239033 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller



























































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)