[English] 日本語
 Yorodumi
Yorodumi- EMDB-17815: Pyrococcus abyssi DNA polymerase D (PolD) in its editing mode bou... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Pyrococcus abyssi DNA polymerase D (PolD) in its editing mode bound to a primer/template substrate containing a mismatch | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Polymerase / DNA / Replication / PolD / Archaea / Editing / Proofreading / Exonuclease / Nuclease / DNA BINDING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationexodeoxyribonuclease I / DNA polymerase complex / single-stranded DNA 3'-5' DNA exonuclease activity / DNA catabolic process / DNA strand elongation involved in DNA replication / DNA polymerase processivity factor activity / leading strand elongation / regulation of DNA replication / DNA-directed DNA polymerase / DNA-directed DNA polymerase activity / DNA binding Similarity search - Function | |||||||||
| Biological species |   Pyrococcus abyssi GE5 (archaea) Pyrococcus abyssi GE5 (archaea) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | |||||||||
 Authors Authors | Betancurt-Anzola L / Martinez-Carranza M / Zatopek KM / Gardner AF / Sauguet L | |||||||||
| Funding support |  France, 1 items France, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Molecular basis for proofreading by the unique exonuclease domain of Family-D DNA polymerases. Authors: Leonardo Betancurt-Anzola / Markel Martínez-Carranza / Marc Delarue / Kelly M Zatopek / Andrew F Gardner / Ludovic Sauguet /   Abstract: Replicative DNA polymerases duplicate entire genomes at high fidelity. This feature is shared among the three domains of life and is facilitated by their dual polymerase and exonuclease activities. ...Replicative DNA polymerases duplicate entire genomes at high fidelity. This feature is shared among the three domains of life and is facilitated by their dual polymerase and exonuclease activities. Family D replicative DNA polymerases (PolD), found exclusively in Archaea, contain an unusual RNA polymerase-like catalytic core, and a unique Mre11-like proofreading active site. Here, we present cryo-EM structures of PolD trapped in a proofreading mode, revealing an unanticipated correction mechanism that extends the repertoire of protein domains known to be involved in DNA proofreading. Based on our experimental structures, mutants of PolD were designed and their contribution to mismatch bypass and exonuclease kinetics was determined. This study sheds light on the convergent evolution of structurally distinct families of DNA polymerases, and the domain acquisition and exchange mechanism that occurred during the evolution of the replisome in the three domains of life. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17815.map.gz emd_17815.map.gz | 230.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17815-v30.xml emd-17815-v30.xml emd-17815.xml emd-17815.xml | 22.8 KB 22.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_17815_fsc.xml emd_17815_fsc.xml | 13.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_17815.png emd_17815.png | 148.4 KB | ||
| Masks |  emd_17815_msk_1.map emd_17815_msk_1.map | 244.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-17815.cif.gz emd-17815.cif.gz | 8 KB | ||
| Others |  emd_17815_half_map_1.map.gz emd_17815_half_map_1.map.gz emd_17815_half_map_2.map.gz emd_17815_half_map_2.map.gz | 226.8 MB 226.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17815 http://ftp.pdbj.org/pub/emdb/structures/EMD-17815 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17815 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17815 | HTTPS FTP |
-Related structure data
| Related structure data |  8pptMC  8ppuC  8ppvC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_17815.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17815.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.86 Å | ||||||||||||||||||||||||||||||||||||
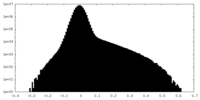
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_17815_msk_1.map emd_17815_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
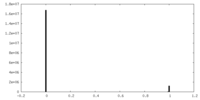
| Density Histograms |
-Half map: #2
| File | emd_17815_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
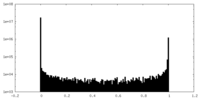
| Density Histograms |
-Half map: #1
| File | emd_17815_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
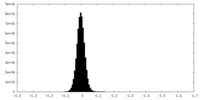
| Density Histograms |
- Sample components
Sample components
-Entire : Binary complex of PolD bound to PCNA and a primer/template duplex...
| Entire | Name: Binary complex of PolD bound to PCNA and a primer/template duplex containing three consecutive mismatches |
|---|---|
| Components |
|
-Supramolecule #1: Binary complex of PolD bound to PCNA and a primer/template duplex...
| Supramolecule | Name: Binary complex of PolD bound to PCNA and a primer/template duplex containing three consecutive mismatches type: complex / ID: 1 / Parent: 0 / Macromolecule list: #3-#4, #1, #5, #2 |
|---|---|
| Source (natural) | Organism:   Pyrococcus abyssi GE5 (archaea) Pyrococcus abyssi GE5 (archaea) |
-Macromolecule #1: DNA polymerase II small subunit
| Macromolecule | Name: DNA polymerase II small subunit / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: DNA-directed DNA polymerase |
|---|---|
| Source (natural) | Organism:   Pyrococcus abyssi GE5 (archaea) Pyrococcus abyssi GE5 (archaea) |
| Molecular weight | Theoretical: 74.009742 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGKHHHHSGH HHTGHHHHSG SHHHTSSSAS TGENLYFQGT GDGSDELVKA LERAGYLLTP SAYYLLVDHF KEGKFSLVEL VKFAKSKGV FIIDGDLAYE FLQFLGLGVP QEIKESYIST GEEAEKTVES QETRASELEE GGVSQVSSGE LQELKEESPE I STTEEEIG ...String: MGKHHHHSGH HHTGHHHHSG SHHHTSSSAS TGENLYFQGT GDGSDELVKA LERAGYLLTP SAYYLLVDHF KEGKFSLVEL VKFAKSKGV FIIDGDLAYE FLQFLGLGVP QEIKESYIST GEEAEKTVES QETRASELEE GGVSQVSSGE LQELKEESPE I STTEEEIG GLELVQSSIS TGSEVEYNNG ENGESVVVLD KYGYPILYAP EEIGEEKEYS KYEDVVIEWN PSVTPVQIEK NY EVKFDVR QVKLRPPKVK NGSGKEGEII VEAYASLFKS RLSKLKRILR ENPEISNVVD IGKLNYVSGD EEVTIIGLVN SKR ETNRGL IFEVEDKTGI VKVFLPKDSE DYREAFKVLP DAVVAFKGFY SKKGIFFANK FYLPDVPLYR KQKPPLEEKV YAIL ISDIH VGSREFCEKA FLKFLEWLNG HVESKEEEEI VSRVKYLIIA GDVVDGIGIY PGQYSDLVIP DIFDQYEALA NLLAN VPEH ITMFIGPGNA DAARPAIPQP EFYKEYAKPI YKLKNAIIIS NPAVIRLHGR DFLIAHGRGI EDVVSFVPGL THHKPG LPM VELLKMRHLA PTFGGKVPIA PDPEDLLVIE EVPDLVQMGH VHVYDAVVYR GVQLVNSATW QAQTEFQKMV NIVPTPA KV PVVDVESARV VKVLDFSGWC UniProtKB: DNA polymerase II small subunit |
-Macromolecule #2: DNA polymerase sliding clamp
| Macromolecule | Name: DNA polymerase sliding clamp / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Pyrococcus abyssi GE5 (archaea) Pyrococcus abyssi GE5 (archaea) |
| Molecular weight | Theoretical: 29.471764 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MRGSHHHHHH GSMPFEIVFE GAKEFAQLIE TASRLIDEAA FKVTEEGISM RAMDPSRVVL IDLNLPASIF SKYEVDGEET IGVNMDHLK KVLKRGKAKE TLILRKGEEN FLEISLQGTA TRTFKLPLID VEEIEVDLPE LPFTAKVVIL GDVIKEAVKD A SLVSDSMK ...String: MRGSHHHHHH GSMPFEIVFE GAKEFAQLIE TASRLIDEAA FKVTEEGISM RAMDPSRVVL IDLNLPASIF SKYEVDGEET IGVNMDHLK KVLKRGKAKE TLILRKGEEN FLEISLQGTA TRTFKLPLID VEEIEVDLPE LPFTAKVVIL GDVIKEAVKD A SLVSDSMK FIAKENEFTM RAEGETQEVE VKLTLEDEGL LDIEVQEETK SAYGISYLSD MVKGLGKADE VTIKFGNEMP MQ MEYYIRD EGRLIFLLAP RVEE UniProtKB: DNA polymerase sliding clamp |
-Macromolecule #5: DP2
| Macromolecule | Name: DP2 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Pyrococcus abyssi GE5 (archaea) Pyrococcus abyssi GE5 (archaea) |
| Molecular weight | Theoretical: 144.418969 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MELPKEMEEY FEMLQREIDK AYEIAKKARA QGKDPSLDVE IPQATDMAGR VESLVGPPGV AKRIRELVKE YGKEIAALKI VDEIIEGKF GDLGSREKYA EQAVRTALAI LTEGIVSAPI EGIANVKIKR NTWADNSEYL ALYYAGPIRS SGGTAQALSV L VGDYVRRK ...String: MELPKEMEEY FEMLQREIDK AYEIAKKARA QGKDPSLDVE IPQATDMAGR VESLVGPPGV AKRIRELVKE YGKEIAALKI VDEIIEGKF GDLGSREKYA EQAVRTALAI LTEGIVSAPI EGIANVKIKR NTWADNSEYL ALYYAGPIRS SGGTAQALSV L VGDYVRRK LGLDRFKPSE KHIERMVEEV DLYHRAVTRL QYHPSPEEVR LAMRNIPIEI TGEATDDVEV SHRDVPGVET NQ LRGGAIL VLAEGVLQKA KKLVKYIDKM GIEGWEWLKE FVEAKEKGEP KEEGKEESLA ESTLEETKVE VDMGFYYSLY QKF KEEIAP SDKYAKEVIG GRPLFSDPSK PGGFRLRYGR SRASGFATWG INPATMILVD EFLAIGTQLK TERPGKGAVV TPVT TIEGP IVKLKDGSVL RVDDYNLALK VREDVEEILY LGDAVIAFGD FVENNQTLLP ANYCEEWWIL EFVKALKEIY EVHLE PFTE NEEESIEEAS DYLEIDPEFL KEMLRDPLRV KPPVELAIHF SEVLGIPLHP YYTLYWNSVE PKDVEKLWRL LKNYAE IEW SNFRGIKFAK KIVISQEKLG DSKRTLELLG LPHTVRDGNV IVDYPWAAAL LTPLGNLNWE FMAKPLYATI DIINENN EI KLRDRGISWI GARMGRPEKA KERKMKPPVQ VLFPIGLAGG SSRDIKKAAE EGKVAEVEIA FFKCPKCGHV GPEHLCPN C GTRKELLWVC PRCNAEYPES QAEGYNYTCP KCNVKLRPYA KRKIRPSELL NRAMENVKVY GVDKLKGVMG MTSGWKMPE PLEKGLLRAK NDVYVFKDGT IRFDATDAPI THFRPREIGV SVEKLRELGY THDFEGKPLV SEDQIVELKP QDIILSKEAG RYLLKVAKF VDDLLEKFYG LPRFYNAEKM EDLIGHLVIG LAPHTSAGIV GRIIGFVDAL VGYAHPYFHA AKRRNCDGDE D AVMLLLDA LLNFSRYYLP EKRGGKMDAP LVITTRLDPR EVDSEVHNMD IVRYYPLEFY EATYELKSPK ELVGVIERVE DR LGKPEMY YGLKFTHDTD DIALGPKMSL YKQLGDMEEK VRRQLEVAKR IRAVDEHGVA EKILNSHLIP DLRGNLRSFT RQE FRCVKC NTKFRRPPLN GKCPVCGGKI VLTVSKGAIE KYLGTAKMLV TEYNVKNYTR QRICLTERDI DSLFENVFPE TQLT LIVNP NDICQRLVMA RTGEVNKSGL LENLSNGSKK TEKAEKAEKP RKKSDEKPKK KRVISLEEFF SRKSK |
-Macromolecule #3: DNA (5'-D(P*CP*CP*GP*GP*GP*CP*CP*GP*AP*GP*CP*CP*GP*TP*GP*CP*TP*TP...
| Macromolecule | Name: DNA (5'-D(P*CP*CP*GP*GP*GP*CP*CP*GP*AP*GP*CP*CP*GP*TP*GP*CP*TP*TP*T)-3') type: dna / ID: 3 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:   Pyrococcus abyssi GE5 (archaea) Pyrococcus abyssi GE5 (archaea) |
| Molecular weight | Theoretical: 5.519542 KDa |
| Sequence | String: (DC)(DG)(DC)(DC)(DG)(DG)(DG)(DC)(DC)(DG) (DA)(DG)(DC)(DC)(DG)(DT)(DG)(DC) |
-Macromolecule #4: DNA (5'-D(P*AP*GP*CP*AP*CP*GP*GP*CP*TP*CP*GP*GP*CP*CP*CP*GP*G)-3')
| Macromolecule | Name: DNA (5'-D(P*AP*GP*CP*AP*CP*GP*GP*CP*TP*CP*GP*GP*CP*CP*CP*GP*G)-3') type: dna / ID: 4 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:   Pyrococcus abyssi GE5 (archaea) Pyrococcus abyssi GE5 (archaea) |
| Molecular weight | Theoretical: 7.741958 KDa |
| Sequence | String: (DA)(DG)(DG)(DT)(DC)(DG)(DT)(DG)(DA)(DA) (DC)(DG)(DG)(DC)(DT)(DC)(DG)(DG)(DC)(DC) (DC)(DG)(DG)(DC)(DG) |
-Macromolecule #6: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 6 / Number of copies: 3 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #7: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 7 / Number of copies: 3 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)