[English] 日本語
 Yorodumi
Yorodumi- EMDB-17756: Structure of the murine trace amine-associated receptor TAAR7f bo... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the murine trace amine-associated receptor TAAR7f bound to N,N-dimethylcyclohexylamine (DMCH) in complex with mini-Gs trimeric G protein | |||||||||
 Map data Map data | Consensus map of the mTAAR7f receptor bound to NDMCH in complex with miniGs trimer | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | trace-amine associated receptor / TAAR / mTAAR7f / GPCR / receptor / G protein / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationtrace-amine receptor activity / sensory perception of chemical stimulus / Olfactory Signaling Pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / G-protein activation / Activation of G protein gated Potassium channels ...trace-amine receptor activity / sensory perception of chemical stimulus / Olfactory Signaling Pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / G-protein activation / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / Prostacyclin signalling through prostacyclin receptor / G beta:gamma signalling through CDC42 / Glucagon signaling in metabolic regulation / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / ADP signalling through P2Y purinoceptor 12 / photoreceptor disc membrane / Sensory perception of sweet, bitter, and umami (glutamate) taste / Glucagon-type ligand receptors / Adrenaline,noradrenaline inhibits insulin secretion / Vasopressin regulates renal water homeostasis via Aquaporins / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / G alpha (z) signalling events / ADP signalling through P2Y purinoceptor 1 / cellular response to catecholamine stimulus / ADORA2B mediated anti-inflammatory cytokines production / G beta:gamma signalling through PI3Kgamma / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / adenylate cyclase-activating dopamine receptor signaling pathway / GPER1 signaling / Inactivation, recovery and regulation of the phototransduction cascade / cellular response to prostaglandin E stimulus / G-protein beta-subunit binding / heterotrimeric G-protein complex / G alpha (12/13) signalling events / sensory perception of taste / extracellular vesicle / signaling receptor complex adaptor activity / Thrombin signalling through proteinase activated receptors (PARs) / retina development in camera-type eye / GTPase binding / Ca2+ pathway / fibroblast proliferation / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / G alpha (i) signalling events / G alpha (s) signalling events / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (q) signalling events / Ras protein signal transduction / Extra-nuclear estrogen signaling / cell population proliferation / G protein-coupled receptor signaling pathway / lysosomal membrane / GTPase activity / synapse / protein-containing complex binding / signal transduction / extracellular exosome / membrane / plasma membrane / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /   | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.92 Å | |||||||||
 Authors Authors | Gusach A / Lee Y / Edwards PC / Huang F / Weyand SN / Tate CG | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Biorxiv / Year: 2023 Journal: Biorxiv / Year: 2023Title: Molecular recognition of an aversive odorant by the murine trace amine-associated receptor TAAR7f. Authors: Gusach A / Lee Y / Khoshgrudi AN / Mukhaleva E / Ma N / Koers EJ / Chen Q / Edwards PC / Huang F / Kim J / Mancia F / Verprintsev DB / Vaidehi N / Weyand SN / Tate CG | |||||||||
| History |
|
- Structure visualization
Structure visualization
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17756.map.gz emd_17756.map.gz | 93 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17756-v30.xml emd-17756-v30.xml emd-17756.xml emd-17756.xml | 37.5 KB 37.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_17756_fsc.xml emd_17756_fsc.xml | 12.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_17756.png emd_17756.png | 121 KB | ||
| Masks |  emd_17756_msk_1.map emd_17756_msk_1.map emd_17756_msk_2.map emd_17756_msk_2.map | 196.4 MB 196.4 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-17756.cif.gz emd-17756.cif.gz | 9.4 KB | ||
| Others |  emd_17756_additional_1.map.gz emd_17756_additional_1.map.gz emd_17756_half_map_1.map.gz emd_17756_half_map_1.map.gz emd_17756_half_map_2.map.gz emd_17756_half_map_2.map.gz | 93.6 MB 182.4 MB 182.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17756 http://ftp.pdbj.org/pub/emdb/structures/EMD-17756 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17756 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17756 | HTTPS FTP |
-Validation report
| Summary document |  emd_17756_validation.pdf.gz emd_17756_validation.pdf.gz | 937 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_17756_full_validation.pdf.gz emd_17756_full_validation.pdf.gz | 936.8 KB | Display | |
| Data in XML |  emd_17756_validation.xml.gz emd_17756_validation.xml.gz | 21.2 KB | Display | |
| Data in CIF |  emd_17756_validation.cif.gz emd_17756_validation.cif.gz | 27.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17756 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17756 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17756 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17756 | HTTPS FTP |
-Related structure data
| Related structure data |  8pm2MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_17756.map.gz / Format: CCP4 / Size: 196.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17756.map.gz / Format: CCP4 / Size: 196.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Consensus map of the mTAAR7f receptor bound to NDMCH in complex with miniGs trimer | ||||||||||||||||||||
| Voxel size | X=Y=Z: 0.824 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : A complex of mouse trace-amine associated receptor 7f solubilized...
| Entire | Name: A complex of mouse trace-amine associated receptor 7f solubilized in LMNG/CHS bound to N,N-dimethylcyclohexylamine and coupled to: Engineered guanine nucleotide-binding protein G(s) subunit ...Name: A complex of mouse trace-amine associated receptor 7f solubilized in LMNG/CHS bound to N,N-dimethylcyclohexylamine and coupled to: Engineered guanine nucleotide-binding protein G(s) subunit alpha isoform short + Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 + Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 + Nanobody 35 |
|---|---|
| Components |
|
-Supramolecule #1: A complex of mouse trace-amine associated receptor 7f solubilized...
| Supramolecule | Name: A complex of mouse trace-amine associated receptor 7f solubilized in LMNG/CHS bound to N,N-dimethylcyclohexylamine and coupled to: Engineered guanine nucleotide-binding protein G(s) subunit ...Name: A complex of mouse trace-amine associated receptor 7f solubilized in LMNG/CHS bound to N,N-dimethylcyclohexylamine and coupled to: Engineered guanine nucleotide-binding protein G(s) subunit alpha isoform short + Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 + Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 + Nanobody 35 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short
| Macromolecule | Name: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 28.964734 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GNSKTEDQRN EEKAQREANK KIEKQLQKDK QVYRATHRLL LLGADNSGKS TIVKQMRILH GGSGGSGGTS GIFETKFQVD KVNFHMFDV GGQRDERRKW IQCFNDVTAI IFVVDSSDYN RLQEALNLFK SIWNNRWLRT ISVILFLNKQ DLLAEKVLAG K SKLEDYFP ...String: GNSKTEDQRN EEKAQREANK KIEKQLQKDK QVYRATHRLL LLGADNSGKS TIVKQMRILH GGSGGSGGTS GIFETKFQVD KVNFHMFDV GGQRDERRKW IQCFNDVTAI IFVVDSSDYN RLQEALNLFK SIWNNRWLRT ISVILFLNKQ DLLAEKVLAG K SKLEDYFP EFARYTTPED ATPEPGEDPR VTRAKYFIRD EFLRISTASG DGRHYCYPHF TCAVDTENAR RIFNDCRDII QR MHLRQYE LL |
-Macromolecule #2: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 37.342785 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: GSELDQLRQE AEQLKNQIRD ARKACADATL SQITNNIDPV GRIQMRTRRT LRGHLAKIYA MHWGTDSRLL VSASQDGKLI IWDSYTTNK VHAIPLRSSW VMTCAYAPSG NYVACGGLDN ICSIYNLKTR EGNVRVSREL AGHTGYLSCC RFLDDNQIVT S SGDTTCAL ...String: GSELDQLRQE AEQLKNQIRD ARKACADATL SQITNNIDPV GRIQMRTRRT LRGHLAKIYA MHWGTDSRLL VSASQDGKLI IWDSYTTNK VHAIPLRSSW VMTCAYAPSG NYVACGGLDN ICSIYNLKTR EGNVRVSREL AGHTGYLSCC RFLDDNQIVT S SGDTTCAL WDIETGQQTT TFTGHTGDVM SLSLAPDTRL FVSGACDASA KLWDVREGMC RQTFTGHESD INAICFFPNG NA FATGSDD ATCRLFDLRA DQELMTYSHD NIICGITSVS FSKSGRLLLA GYDDFNCNVW DALKADRAGV LAGHDNRVSC LGV TDDGMA VATGSWDSFL KIWN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 3 / Details: There is an engineered mutation C68S introduced / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.845078 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFSAI L UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #4: Nanobody 35
| Macromolecule | Name: Nanobody 35 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 16.926076 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKYLLPTAAA GLLLLAAQPA MAQVQLQESG GGLVQPGGSL RLSCAASGFT FSNYKMNWVR QAPGKGLEWV SDISQSGASI SYTGSVKGR FTISRDNAKN TLYLQMNSLK PEDTAVYYCA RCPAPFTRDC FDVTSTTYAY RGQGTQVTVS SHHHHHH |
-Macromolecule #5: Trace amine-associated receptor 7f
| Macromolecule | Name: Trace amine-associated receptor 7f / type: protein_or_peptide / ID: 5 Details: mTAAR7f sequence contains cleaved protease sites: TEV (S residue on the N terminus) and HRV-3C (C terminus) Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 41.012539 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: SMSIADETVS WNQDSILSRD LFSATSAELC YENLNRSCVR SPYSPGPRLI LYAVFGFGAV LAVCGNLLVM TSILHFRQLH SPANFLVAS LACADFLVGV MVMPFSMVRS VEGCWYFGDS YCKLHTCFDV SFCYCSLFHL CFISVDRYIA VSDPLAYPTR F TASVSGKC ...String: SMSIADETVS WNQDSILSRD LFSATSAELC YENLNRSCVR SPYSPGPRLI LYAVFGFGAV LAVCGNLLVM TSILHFRQLH SPANFLVAS LACADFLVGV MVMPFSMVRS VEGCWYFGDS YCKLHTCFDV SFCYCSLFHL CFISVDRYIA VSDPLAYPTR F TASVSGKC ITFSWLLSIS YGFSLIYTGA SEAGLEDLVS SLTCVGGCQI AVNQTWVFIN FSVFLIPTLV MITVYSKIFL IA KQQAQNI EKMSKQTARA SDSYKDRVAK RERKAAKTLG IAVAAFLLSW LPYFIDSFID AFLGFITPTY VYEILVWIVY YNS AMNPLI YAFFYPWFRK AIKLTVTGKI LRENSSTTNL FSELEVLFQ UniProtKB: Trace amine-associated receptor 7f |
-Macromolecule #6: CHOLESTEROL HEMISUCCINATE
| Macromolecule | Name: CHOLESTEROL HEMISUCCINATE / type: ligand / ID: 6 / Number of copies: 1 / Formula: Y01 |
|---|---|
| Molecular weight | Theoretical: 486.726 Da |
| Chemical component information |  ChemComp-Y01: |
-Macromolecule #7: ~{N},~{N}-dimethylcyclohexanamine
| Macromolecule | Name: ~{N},~{N}-dimethylcyclohexanamine / type: ligand / ID: 7 / Number of copies: 1 / Formula: 8IA |
|---|---|
| Molecular weight | Theoretical: 127.227 Da |
| Chemical component information | 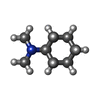 ChemComp-8IA: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.8 mg/mL | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||||||||||||||
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 400 / Support film - Material: GOLD / Support film - topology: HOLEY ARRAY / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 120 sec. / Pretreatment - Atmosphere: OTHER Details: Forward Power of 38 W, Reflected Power of 2W; Fischione | ||||||||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Number grids imaged: 1 / Number real images: 11157 / Average exposure time: 7.84 sec. / Average electron dose: 55.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Calibrated defocus max: 2.4 µm / Calibrated defocus min: 2.4 µm / Calibrated magnification: 96000 / Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.4 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 96000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | Chain - Source name: Other / Chain - Initial model type: other Details: Initial models of heterotrimeric mini-Gs and Nb35 were sourced from PDB 7T9I. A de novo model of TAAR7f was generated from the focused map and protein sequence using ModelAngelo |
|---|---|
| Refinement | Space: REAL / Protocol: OTHER |
| Output model |  PDB-8pm2: |
 Movie
Movie Controller
Controller























