[English] 日本語
 Yorodumi
Yorodumi- EMDB-17685: E. coli transcription complex paused at ops site and bound to Rfa... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | E. coli transcription complex paused at ops site and bound to RfaH and NusA | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | transcription complex / ops / RfaH / NusA / TRANSCRIPTION | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationregulatory RNA binding / transcription antitermination factor activity, DNA binding / translation activator activity / DNA-templated transcription elongation / transcription elongation-coupled chromatin remodeling / RNA polymerase complex / submerged biofilm formation / cellular response to cell envelope stress / regulation of DNA-templated transcription initiation / protein complex oligomerization ...regulatory RNA binding / transcription antitermination factor activity, DNA binding / translation activator activity / DNA-templated transcription elongation / transcription elongation-coupled chromatin remodeling / RNA polymerase complex / submerged biofilm formation / cellular response to cell envelope stress / regulation of DNA-templated transcription initiation / protein complex oligomerization / bacterial-type flagellum assembly / bacterial-type RNA polymerase core enzyme binding / cytosolic DNA-directed RNA polymerase complex / bacterial-type flagellum-dependent cell motility / nitrate assimilation / regulation of DNA-templated transcription elongation / transcription elongation factor complex / positive regulation of translation / transcription antitermination / DNA-directed RNA polymerase complex / cell motility / DNA-templated transcription initiation / DNA-templated transcription termination / ribonucleoside binding / DNA-directed RNA polymerase / DNA-directed RNA polymerase activity / ribosome biogenesis / response to heat / protein-containing complex assembly / intracellular iron ion homeostasis / protein dimerization activity / DNA-binding transcription factor activity / protein domain specific binding / response to antibiotic / nucleotide binding / DNA-templated transcription / magnesium ion binding / DNA binding / RNA binding / zinc ion binding / membrane / cytoplasm / cytosol Similarity search - Function | ||||||||||||
| Biological species |  | ||||||||||||
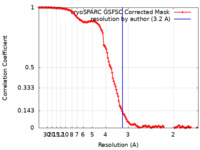
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | ||||||||||||
 Authors Authors | Zuber PK / Said N / Hilal T / Loll B / Wahl MC / Knauer SH | ||||||||||||
| Funding support |  Germany, 3 items Germany, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Concerted transformation of a hyper-paused transcription complex and its reinforcing protein. Authors: Philipp K Zuber / Nelly Said / Tarek Hilal / Bing Wang / Bernhard Loll / Jorge González-Higueras / César A Ramírez-Sarmiento / Georgiy A Belogurov / Irina Artsimovitch / Markus C Wahl / Stefan H Knauer /      Abstract: RfaH, a paralog of the universally conserved NusG, binds to RNA polymerases (RNAP) and ribosomes to activate expression of virulence genes. In free, autoinhibited RfaH, an α-helical KOW domain ...RfaH, a paralog of the universally conserved NusG, binds to RNA polymerases (RNAP) and ribosomes to activate expression of virulence genes. In free, autoinhibited RfaH, an α-helical KOW domain sequesters the RNAP-binding site. Upon recruitment to RNAP paused at an ops site, KOW is released and refolds into a β-barrel, which binds the ribosome. Here, we report structures of ops-paused transcription elongation complexes alone and bound to the autoinhibited and activated RfaH, which reveal swiveled, pre-translocated pause states stabilized by an ops hairpin in the non-template DNA. Autoinhibited RfaH binds and twists the ops hairpin, expanding the RNA:DNA hybrid to 11 base pairs and triggering the KOW release. Once activated, RfaH hyper-stabilizes the pause, which thus requires anti-backtracking factors for escape. Our results suggest that the entire RfaH cycle is solely determined by the ops and RfaH sequences and provide insights into mechanisms of recruitment and metamorphosis of NusG homologs across all life. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17685.map.gz emd_17685.map.gz | 54.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17685-v30.xml emd-17685-v30.xml emd-17685.xml emd-17685.xml | 32.4 KB 32.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_17685_fsc.xml emd_17685_fsc.xml | 14.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_17685.png emd_17685.png | 99 KB | ||
| Filedesc metadata |  emd-17685.cif.gz emd-17685.cif.gz | 9.7 KB | ||
| Others |  emd_17685_half_map_1.map.gz emd_17685_half_map_1.map.gz emd_17685_half_map_2.map.gz emd_17685_half_map_2.map.gz | 200.8 MB 200.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17685 http://ftp.pdbj.org/pub/emdb/structures/EMD-17685 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17685 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17685 | HTTPS FTP |
-Related structure data
| Related structure data |  8pilMC  8pdyC  8penC  8pfgC  8pfjC  8ph9C  8phkC  8pibC  8pidC  8pimC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_17685.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17685.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.832 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_17685_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_17685_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : transcription complex paused at ops site, assembled on a fully co...
+Supramolecule #1: transcription complex paused at ops site, assembled on a fully co...
+Supramolecule #2: Transcription complex
+Supramolecule #3: DNA and RNA
+Macromolecule #1: Transcription antitermination protein RfaH
+Macromolecule #2: DNA-directed RNA polymerase subunit beta
+Macromolecule #3: DNA-directed RNA polymerase subunit beta'
+Macromolecule #4: DNA-directed RNA polymerase subunit omega
+Macromolecule #5: DNA-directed RNA polymerase subunit alpha
+Macromolecule #9: Transcription termination/antitermination protein NusA
+Macromolecule #6: non-template DNA
+Macromolecule #7: template DNA
+Macromolecule #8: RNA
+Macromolecule #10: ZINC ION
+Macromolecule #11: MAGNESIUM ION
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3.3 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 283 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 3142 / Average exposure time: 40.57 sec. / Average electron dose: 42.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 96000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Overall B value: 121 |
|---|---|
| Output model |  PDB-8pil: |
 Movie
Movie Controller
Controller





















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)