[English] 日本語
 Yorodumi
Yorodumi- EMDB-17629: Symmetry expanded D7 local refined map of mitochondrial heat-shoc... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Symmetry expanded D7 local refined map of mitochondrial heat-shock protein 60-like protein from Chaetomium thermophilum | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | mitochondrial heat-shock protein 60-like protein / Stress response / ATP-binding / Nucleotide-binding / Chaetomium thermophilum / native cell extracts / CHAPERONE | |||||||||
| Function / homology |  Function and homology information Function and homology informationresponse to stress / ATP-dependent protein folding chaperone / protein refolding / ATP binding Similarity search - Function | |||||||||
| Biological species |  Thermochaetoides thermophila (fungus) Thermochaetoides thermophila (fungus) | |||||||||
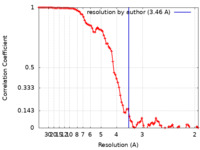
| Method | single particle reconstruction / cryo EM / Resolution: 3.46 Å | |||||||||
 Authors Authors | Semchonok DA / Kyrilis FL / Hamdi F / Kastritis PL | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Symmetry expanded D7, locally refined cryo-EM map of mitochondrial heat shock protein 60-like protein Chaetomium thermophilum. Authors: Semchonok DA / Kyrilis FL / Hamdi F / Kastritis PL | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17629.map.gz emd_17629.map.gz | 59.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17629-v30.xml emd-17629-v30.xml emd-17629.xml emd-17629.xml | 16.4 KB 16.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_17629_fsc.xml emd_17629_fsc.xml | 8.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_17629.png emd_17629.png | 84.1 KB | ||
| Masks |  emd_17629_msk_1.map emd_17629_msk_1.map | 64 MB |  Mask map Mask map | |
| Others |  emd_17629_half_map_1.map.gz emd_17629_half_map_1.map.gz emd_17629_half_map_2.map.gz emd_17629_half_map_2.map.gz | 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17629 http://ftp.pdbj.org/pub/emdb/structures/EMD-17629 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17629 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17629 | HTTPS FTP |
-Validation report
| Summary document |  emd_17629_validation.pdf.gz emd_17629_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_17629_full_validation.pdf.gz emd_17629_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_17629_validation.xml.gz emd_17629_validation.xml.gz | 16.2 KB | Display | |
| Data in CIF |  emd_17629_validation.cif.gz emd_17629_validation.cif.gz | 21.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17629 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17629 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17629 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17629 | HTTPS FTP |
-Related structure data
| Related structure data |  8pe8MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_17629.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17629.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.96 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_17629_msk_1.map emd_17629_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_17629_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_17629_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : native cell extract of Chaetomium thermophilum
| Entire | Name: native cell extract of Chaetomium thermophilum |
|---|---|
| Components |
|
-Supramolecule #1: native cell extract of Chaetomium thermophilum
| Supramolecule | Name: native cell extract of Chaetomium thermophilum / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: all Details: Purified and fractionated (fraction 25) cell lysate of Chaetomium thermophilum |
|---|---|
| Source (natural) | Organism:  Thermochaetoides thermophila (fungus) Thermochaetoides thermophila (fungus) |
| Molecular weight | Theoretical: 60 KDa |
-Macromolecule #1: Mitochondrial heat shock protein 60-like protein
| Macromolecule | Name: Mitochondrial heat shock protein 60-like protein / type: protein_or_peptide / ID: 1 / Number of copies: 14 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Thermochaetoides thermophila (fungus) Thermochaetoides thermophila (fungus) |
| Molecular weight | Theoretical: 60.669473 KDa |
| Sequence | String: MQRALTRASV SSPLATARVR SAQQLRFAHK ELKFGVEGRA ALLNGVETLA KAVATTLGPK GRNVLIESTF GSPKITKDGV TVAKAISLK DKFENLGAKL LAEVASKTNE VAGDGTTTAT VLARAIFSEM VKNVAAGCNP MDLRRGIQAA VDAVVEYLQQ N KRDITTSA ...String: MQRALTRASV SSPLATARVR SAQQLRFAHK ELKFGVEGRA ALLNGVETLA KAVATTLGPK GRNVLIESTF GSPKITKDGV TVAKAISLK DKFENLGAKL LAEVASKTNE VAGDGTTTAT VLARAIFSEM VKNVAAGCNP MDLRRGIQAA VDAVVEYLQQ N KRDITTSA EIAQVATISA NGDQHIGKLI ASAMEKVGKE GVITVKEGKT LQDELEVTEG MRFDRGFVSP YFITDAKAQK VE FEKPLIL LSEQKISAAT DIIPALEISH KMRRPLVIIA EDIDGEALAV CILNKLRGQL EVAAVKAPGF GDNRKSILGD IAV LTNGTV FTNELDVKLE KVTPDMLGST GSITITKEDT IILNGDGSKD AIAQRCEQIR GAMNDPSTSE YEKEKLQERL AKLS GGVAV IKVGGSSEVE VGEKKDRFVD ALNATRAAVE EGILPGGGTA LIKASVNALN NLKPANFDQQ LGVNIIKNAI TRPAR MIVE NAGLEGSVVI GKISDEYAAD FNKGFNSATG EYVDMIQAGI LDPLKVVRTG LIDASGVASL LGTTEVAIVE APEEKG PAG GMGGMGGMGG MM UniProtKB: Mitochondrial heat shock protein 60-like protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.3 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Specialist optics | Phase plate: OTHER |
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 1109 / Average electron dose: 28.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm |
| Sample stage | Cooling holder cryogen: NITROGEN |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Chain ID: A / Chain - Source name: AlphaFold / Chain - Initial model type: in silico model Details: The initial model consisted of the complete monomeric unit of HSP60 |
|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
| Output model |  PDB-8pe8: |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)