+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | cryo-EM structure of Doa10 in MSP1E3D1 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ERAD / Doa10 / March6 / TEB4 / retrotranslocation / ubiquitination / Ubc6 / sybody / SQLE / squalenemonooxygenase / LIGASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationDoa10p ubiquitin ligase complex / nuclear inner membrane / retrograde protein transport, ER to cytosol / ERAD pathway / RING-type E3 ubiquitin transferase / ubiquitin-protein transferase activity / ubiquitin protein ligase activity / nuclear envelope / protein ubiquitination / endoplasmic reticulum membrane ...Doa10p ubiquitin ligase complex / nuclear inner membrane / retrograde protein transport, ER to cytosol / ERAD pathway / RING-type E3 ubiquitin transferase / ubiquitin-protein transferase activity / ubiquitin protein ligase activity / nuclear envelope / protein ubiquitination / endoplasmic reticulum membrane / endoplasmic reticulum / zinc ion binding / membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
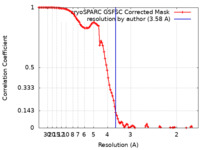
| Method | single particle reconstruction / cryo EM / Resolution: 3.58 Å | |||||||||
 Authors Authors | Botsch JJ / Braeuning B / Schulman BA | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Doa10/MARCH6 architecture interconnects E3 ligase activity with lipid-binding transmembrane channel to regulate SQLE. Authors: J Josephine Botsch / Roswitha Junker / Michèle Sorgenfrei / Patricia P Ogger / Luca Stier / Susanne von Gronau / Peter J Murray / Markus A Seeger / Brenda A Schulman / Bastian Bräuning /   Abstract: Transmembrane E3 ligases play crucial roles in homeostasis. Much protein and organelle quality control, and metabolic regulation, are determined by ER-resident MARCH6 E3 ligases, including Doa10 in ...Transmembrane E3 ligases play crucial roles in homeostasis. Much protein and organelle quality control, and metabolic regulation, are determined by ER-resident MARCH6 E3 ligases, including Doa10 in yeast. Here, we present Doa10/MARCH6 structural analysis by cryo-EM and AlphaFold predictions, and a structure-based mutagenesis campaign. The majority of Doa10/MARCH6 adopts a unique circular structure within the membrane. This channel is established by a lipid-binding scaffold, and gated by a flexible helical bundle. The ubiquitylation active site is positioned over the channel by connections between the cytosolic E3 ligase RING domain and the membrane-spanning scaffold and gate. Here, by assaying 95 MARCH6 variants for effects on stability of the well-characterized substrate SQLE, which regulates cholesterol levels, we reveal crucial roles of the gated channel and RING domain consistent with AlphaFold-models of substrate-engaged and ubiquitylation complexes. SQLE degradation further depends on connections between the channel and RING domain, and lipid binding sites, revealing how interconnected Doa10/MARCH6 elements could orchestrate metabolic signals, substrate binding, and E3 ligase activity. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17597.map.gz emd_17597.map.gz | 101.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17597-v30.xml emd-17597-v30.xml emd-17597.xml emd-17597.xml | 15.9 KB 15.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_17597_fsc.xml emd_17597_fsc.xml | 10.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_17597.png emd_17597.png | 88.4 KB | ||
| Masks |  emd_17597_msk_1.map emd_17597_msk_1.map | 107.2 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-17597.cif.gz emd-17597.cif.gz | 6.7 KB | ||
| Others |  emd_17597_half_map_1.map.gz emd_17597_half_map_1.map.gz emd_17597_half_map_2.map.gz emd_17597_half_map_2.map.gz | 99.6 MB 99.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17597 http://ftp.pdbj.org/pub/emdb/structures/EMD-17597 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17597 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17597 | HTTPS FTP |
-Related structure data
| Related structure data |  8pd0MC  8pdaC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_17597.map.gz / Format: CCP4 / Size: 107.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17597.map.gz / Format: CCP4 / Size: 107.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.8512 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_17597_msk_1.map emd_17597_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_17597_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_17597_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Doa10 with Ubc6 and sybody in MSP1E3D1
| Entire | Name: Doa10 with Ubc6 and sybody in MSP1E3D1 |
|---|---|
| Components |
|
-Supramolecule #1: Doa10 with Ubc6 and sybody in MSP1E3D1
| Supramolecule | Name: Doa10 with Ubc6 and sybody in MSP1E3D1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 150 KDa |
-Macromolecule #1: ERAD-associated E3 ubiquitin-protein ligase DOA10
| Macromolecule | Name: ERAD-associated E3 ubiquitin-protein ligase DOA10 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: RING-type E3 ubiquitin transferase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 151.608109 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MDVDSDVNVS RLRDELHKVA NEETDTATFN DDAPSGATCR ICRGEATEDN PLFHPCKCRG SIKYMHESCL LEWVASKNID ISKPGADVK CDICHYPIQF KTIYAENMPE KIPFSLLLSK SILTFFEKAR LALTIGLAAV LYIIGVPLVW NMFGKLYTMM L DGSSPYPG ...String: MDVDSDVNVS RLRDELHKVA NEETDTATFN DDAPSGATCR ICRGEATEDN PLFHPCKCRG SIKYMHESCL LEWVASKNID ISKPGADVK CDICHYPIQF KTIYAENMPE KIPFSLLLSK SILTFFEKAR LALTIGLAAV LYIIGVPLVW NMFGKLYTMM L DGSSPYPG DFLKSLIYGY DQSATPELTT RAIFYQLLQN HSFTSLQFIM IVILHIALYF QYDMIVREDV FSKMVFHKIG PR LSPKDLK SRLKERFPMM DDRMVEYLAR EMRAHDENRQ EQGHDRLNMP AAAADNNNNV INPRNDNVPP QDPNDHRNFE NLR HVDELD HDEATEEHEN NDSDNSLPSG DDSSRILPGS SSDNEEDEEA EGQQQQQQPE EEADYRDHIE PNPIDMWANR RAQN EFDDL IAAQQNAINR PNAPVFIPPP AQNRAGNVDQ DEQDFGAAVG VPPAQANPDD QGQGPLVINL KLKLLNVIAY FIIAV VFTA IYLAISYLFP TFIGFGLLKI YFGIFKVILR GLCHLYYLSG AHIAYNGLTK LVPKVDVAMS WISDHLIHDI IYLYNG YTE NTMKHSIFIR ALPALTTYLT SVSIVCASSN LVSRGYGREN GMSNPTRRLI FQILFALKCT FKVFTLFFIE LAGFPIL AG VMLDFSLFCP ILASNSRMLW VPSICAIWPP FSLFVYWTIG TLYMYWFAKY IGMIRKNIIR PGVLFFIRSP EDPNIKIL H DSLIHPMSIQ LSRLCLSMFI YAIFIVLGFG FHTRIFFPFM LKSNLLSVPE AYKPTSIISW KFNTILLTLY FTKRILESS SYVKPLLERY WKTIFKLCSR KLRLSSFILG KDTPTERGHI VYRNLFYKYI AAKNAEWSNQ ELFTKPKTLE QAEELFGQVR DVHAYFVPD GVLMRVPSSD IVSRNYVQTM FVPVTKDDKL LKPLDLERIK ERNKRAAGEF GYLDEQNTEY DQYYIVYVPP D FRLRYMTL LGLVWLFASI LMLGVTFISQ ALINFVCSFG FLPVVKLLLG ERNKVYVAWK ELSDISYSYL NIYYVCVGSV CL SKIAKDI LHFTEGQNTL DEHAVDENEV EEVEHDIPER DINNAPVNNI NNVEEGQGIF MAIFNSIFDS MLVKYNLMVF IAI MIAVIR TMVSWVVLTD GILACYNYLT IRVFGNSSYT IGNSKWFKYD ESLLFVVWII SSMVNFGTGY KSLKLFFRNR NTSK LNFLK TMALELFKQG FLHMVIYVLP IIILSLVFLR DVSTKQIIDI SHGSRSFTLS LNESFPTWTR MQDIYFGLLI ALESF TFFF QATVLFIQWF KSTVQNVKDE VYTKGRALEN LPDES UniProtKB: ERAD-associated E3 ubiquitin-protein ligase DOA10 |
-Macromolecule #2: 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE
| Macromolecule | Name: 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE / type: ligand / ID: 2 / Number of copies: 3 / Formula: PCW |
|---|---|
| Molecular weight | Theoretical: 787.121 Da |
| Chemical component information |  ChemComp-PCW: |
-Macromolecule #3: 1,2-DIPALMITOYL-SN-GLYCERO-3-PHOSPHATE
| Macromolecule | Name: 1,2-DIPALMITOYL-SN-GLYCERO-3-PHOSPHATE / type: ligand / ID: 3 / Number of copies: 1 / Formula: PX6 |
|---|---|
| Molecular weight | Theoretical: 647.883 Da |
| Chemical component information |  ChemComp-PX6: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 70.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.8000000000000003 µm / Nominal defocus min: 0.7000000000000001 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | Chain - Source name: AlphaFold / Chain - Initial model type: in silico model |
|---|---|
| Output model |  PDB-8pd0: |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)