+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | Mouse RPL39 integrated into the yeast 60S ribosomal subunit | |||||||||
 マップデータ マップデータ | ||||||||||
 試料 試料 |
| |||||||||
 キーワード キーワード | 60S ribosomal subunit / protein exit tunnel / RPL39 / RPL39L / RIBOSOME | |||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報Formation of a pool of free 40S subunits / SRP-dependent cotranslational protein targeting to membrane / Major pathway of rRNA processing in the nucleolus and cytosol / Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) / Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) / L13a-mediated translational silencing of Ceruloplasmin expression / GTP hydrolysis and joining of the 60S ribosomal subunit / pre-mRNA 5'-splice site binding / response to cycloheximide / cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) ...Formation of a pool of free 40S subunits / SRP-dependent cotranslational protein targeting to membrane / Major pathway of rRNA processing in the nucleolus and cytosol / Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) / Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) / L13a-mediated translational silencing of Ceruloplasmin expression / GTP hydrolysis and joining of the 60S ribosomal subunit / pre-mRNA 5'-splice site binding / response to cycloheximide / cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / SRP-dependent cotranslational protein targeting to membrane / GTP hydrolysis and joining of the 60S ribosomal subunit / negative regulation of mRNA splicing, via spliceosome / Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) / Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) / preribosome, large subunit precursor / Formation of a pool of free 40S subunits / L13a-mediated translational silencing of Ceruloplasmin expression / translational elongation / ribosomal large subunit export from nucleus / protein-RNA complex assembly / regulation of translational fidelity / translational termination / maturation of LSU-rRNA / ribosomal large subunit biogenesis / maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / innate immune response in mucosa / macroautophagy / translational initiation / maintenance of translational fidelity / modification-dependent protein catabolic process / protein tag activity / rRNA processing / antimicrobial humoral immune response mediated by antimicrobial peptide / antibacterial humoral response / ribosome biogenesis / 5S rRNA binding / ribosomal large subunit assembly / large ribosomal subunit rRNA binding / cytosolic large ribosomal subunit / cytoplasmic translation / negative regulation of translation / defense response to Gram-positive bacterium / rRNA binding / protein ubiquitination / structural constituent of ribosome / ribosome / translation / response to antibiotic / mRNA binding / ubiquitin protein ligase binding / nucleolus / extracellular space / RNA binding / zinc ion binding / nucleus / cytosol / cytoplasm 類似検索 - 分子機能 | |||||||||
| 生物種 |   | |||||||||
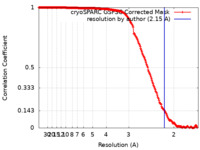
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 2.15 Å | |||||||||
 データ登録者 データ登録者 | Rabl J / Banerjee A / Boehringer D / Zavolan M | |||||||||
| 資金援助 |  スイス, 1件 スイス, 1件
| |||||||||
 引用 引用 | ジャーナル: Acta Crystallogr D Struct Biol / 年: 2018 タイトル: Real-space refinement in PHENIX for cryo-EM and crystallography. 著者: Pavel V Afonine / Billy K Poon / Randy J Read / Oleg V Sobolev / Thomas C Terwilliger / Alexandre Urzhumtsev / Paul D Adams /    要旨: This article describes the implementation of real-space refinement in the phenix.real_space_refine program from the PHENIX suite. The use of a simplified refinement target function enables very fast ...This article describes the implementation of real-space refinement in the phenix.real_space_refine program from the PHENIX suite. The use of a simplified refinement target function enables very fast calculation, which in turn makes it possible to identify optimal data-restraint weights as part of routine refinements with little runtime cost. Refinement of atomic models against low-resolution data benefits from the inclusion of as much additional information as is available. In addition to standard restraints on covalent geometry, phenix.real_space_refine makes use of extra information such as secondary-structure and rotamer-specific restraints, as well as restraints or constraints on internal molecular symmetry. The re-refinement of 385 cryo-EM-derived models available in the Protein Data Bank at resolutions of 6 Å or better shows significant improvement of the models and of the fit of these models to the target maps. | |||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| 添付画像 |
|---|
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_17550.map.gz emd_17550.map.gz | 775.2 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-17550-v30.xml emd-17550-v30.xml emd-17550.xml emd-17550.xml | 62.7 KB 62.7 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| FSC (解像度算出) |  emd_17550_fsc.xml emd_17550_fsc.xml | 19.7 KB | 表示 |  FSCデータファイル FSCデータファイル |
| 画像 |  emd_17550.png emd_17550.png | 149.6 KB | ||
| マスクデータ |  emd_17550_msk_1.map emd_17550_msk_1.map | 824 MB |  マスクマップ マスクマップ | |
| Filedesc metadata |  emd-17550.cif.gz emd-17550.cif.gz | 13.7 KB | ||
| その他 |  emd_17550_half_map_1.map.gz emd_17550_half_map_1.map.gz emd_17550_half_map_2.map.gz emd_17550_half_map_2.map.gz | 764 MB 763.9 MB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17550 http://ftp.pdbj.org/pub/emdb/structures/EMD-17550 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17550 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17550 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_17550_validation.pdf.gz emd_17550_validation.pdf.gz | 1 MB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_17550_full_validation.pdf.gz emd_17550_full_validation.pdf.gz | 1 MB | 表示 | |
| XML形式データ |  emd_17550_validation.xml.gz emd_17550_validation.xml.gz | 28.3 KB | 表示 | |
| CIF形式データ |  emd_17550_validation.cif.gz emd_17550_validation.cif.gz | 37.6 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17550 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17550 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17550 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17550 | HTTPS FTP |
-関連構造データ
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| 「今月の分子」の関連する項目 |
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_17550.map.gz / 形式: CCP4 / 大きさ: 824 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_17550.map.gz / 形式: CCP4 / 大きさ: 824 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 0.84 Å | ||||||||||||||||||||||||||||||||||||
| 密度 |
| ||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
|
-添付データ
-マスク #1
| ファイル |  emd_17550_msk_1.map emd_17550_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
-ハーフマップ: #2
| ファイル | emd_17550_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
-ハーフマップ: #1
| ファイル | emd_17550_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
- 試料の構成要素
試料の構成要素
+全体 : Mouse RPL39 integrated into yeast 60S ribosomal subunit
+超分子 #1: Mouse RPL39 integrated into yeast 60S ribosomal subunit
+分子 #1: Large ribosomal subunit protein eL39
+分子 #2: 60S ribosomal protein L7-A
+分子 #3: 60S ribosomal protein L25
+分子 #4: 60S ribosomal protein L38
+分子 #6: 60S ribosomal protein L8-A
+分子 #7: 60S ribosomal protein L26-A
+分子 #8: Ubiquitin-60S ribosomal protein L40
+分子 #9: 60S ribosomal protein L13-A
+分子 #10: 60S ribosomal protein L9-A
+分子 #11: 60S ribosomal protein L27-A
+分子 #12: 60S ribosomal protein L42-A
+分子 #13: 60S ribosomal protein L14-A
+分子 #14: 60S ribosomal protein L10
+分子 #15: 60S ribosomal protein L28
+分子 #16: 60S ribosomal protein L43-A
+分子 #17: 60S ribosomal protein L15-A
+分子 #18: 60S ribosomal protein L11-A
+分子 #19: 60S ribosomal protein L29
+分子 #20: 60S ribosomal protein L17-A
+分子 #21: 60S ribosomal protein L16-A
+分子 #22: 60S ribosomal protein L18-A
+分子 #23: 60S ribosomal protein L30
+分子 #24: 60S ribosomal protein L19-A
+分子 #26: 60S ribosomal protein L20-A
+分子 #27: 60S ribosomal protein L31-A
+分子 #28: 60S ribosomal protein L21-A
+分子 #30: 60S ribosomal protein L22-A
+分子 #31: 60S ribosomal protein L32
+分子 #32: 60S ribosomal protein L23-A
+分子 #33: 60S ribosomal protein L2-A
+分子 #34: 60S ribosomal protein L24-A
+分子 #35: 60S ribosomal protein L33-A
+分子 #36: 60S ribosomal protein L3
+分子 #37: 60S ribosomal protein L34-A
+分子 #38: 60S ribosomal protein L4-A
+分子 #39: 60S ribosomal protein L35-A
+分子 #40: 60S ribosomal protein L5
+分子 #41: 60S ribosomal protein L36-A
+分子 #42: 60S ribosomal protein L6-A
+分子 #43: 60S ribosomal protein L37-A
+分子 #5: 25S rRNA
+分子 #25: 5.8S rRNA
+分子 #29: 5S rRNA
+分子 #44: MAGNESIUM ION
+分子 #45: CHLORIDE ION
+分子 #46: SPERMIDINE
+分子 #47: SPERMINE
+分子 #48: ZINC ION
+分子 #49: water
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 緩衝液 | pH: 7.6 |
|---|---|
| 凍結 | 凍結剤: ETHANE-PROPANE |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI TITAN KRIOS |
|---|---|
| 撮影 | フィルム・検出器のモデル: GATAN K3 BIOQUANTUM (6k x 4k) 平均電子線量: 45.0 e/Å2 |
| 電子線 | 加速電圧: 300 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD / 最大 デフォーカス(公称値): 2.5 µm / 最小 デフォーカス(公称値): 1.0 µm |
| 実験機器 |  モデル: Titan Krios / 画像提供: FEI Company |
+ 画像解析
画像解析
-原子モデル構築 1
| 精密化 | 空間: REAL / プロトコル: OTHER |
|---|---|
| 得られたモデル |  PDB-8p8n: |
 ムービー
ムービー コントローラー
コントローラー



















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)