[English] 日本語
 Yorodumi
Yorodumi- EMDB-17466: Cryo-EM map of HECT E3 ligase UBR5 forming K48 linked ubiquitin chains -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM map of HECT E3 ligase UBR5 forming K48 linked ubiquitin chains | |||||||||
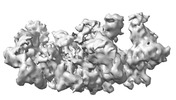
 Map data Map data | DeepEMhancer map of UBR5~UbD~UbA | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | E3 ligase / UBR5 / Ubiquitination / UBQ / Ubiquitin / HECT / K48 / UBQ-chain / polyubiquitylation / LIGASE | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
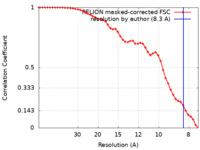
| Method | single particle reconstruction / cryo EM / Resolution: 8.3 Å | |||||||||
 Authors Authors | Hehl LA / Prabu JR / Schulman BA | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Chem Biol / Year: 2024 Journal: Nat Chem Biol / Year: 2024Title: Structural snapshots along K48-linked ubiquitin chain formation by the HECT E3 UBR5. Authors: Laura A Hehl / Daniel Horn-Ghetko / J Rajan Prabu / Ronnald Vollrath / D Tung Vu / David A Pérez Berrocal / Monique P C Mulder / Gerbrand J van der Heden van Noort / Brenda A Schulman /   Abstract: Ubiquitin (Ub) chain formation by homologous to E6AP C-terminus (HECT)-family E3 ligases regulates vast biology, yet the structural mechanisms remain unknown. We used chemistry and cryo-electron ...Ubiquitin (Ub) chain formation by homologous to E6AP C-terminus (HECT)-family E3 ligases regulates vast biology, yet the structural mechanisms remain unknown. We used chemistry and cryo-electron microscopy (cryo-EM) to visualize stable mimics of the intermediates along K48-linked Ub chain formation by the human E3, UBR5. The structural data reveal a ≈ 620 kDa UBR5 dimer as the functional unit, comprising a scaffold with flexibly tethered Ub-associated (UBA) domains, and elaborately arranged HECT domains. Chains are forged by a UBA domain capturing an acceptor Ub, with its K48 lured into the active site by numerous interactions between the acceptor Ub, manifold UBR5 elements and the donor Ub. The cryo-EM reconstructions allow defining conserved HECT domain conformations catalyzing Ub transfer from E2 to E3 and from E3. Our data show how a full-length E3, ubiquitins to be adjoined, E2 and intermediary products guide a feed-forward HECT domain conformational cycle establishing a highly efficient, broadly targeting, K48-linked Ub chain forging machine. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17466.map.gz emd_17466.map.gz | 7.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17466-v30.xml emd-17466-v30.xml emd-17466.xml emd-17466.xml | 14.1 KB 14.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_17466_fsc.xml emd_17466_fsc.xml | 4.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_17466.png emd_17466.png | 61.3 KB | ||
| Filedesc metadata |  emd-17466.cif.gz emd-17466.cif.gz | 4 KB | ||
| Others |  emd_17466_half_map_1.map.gz emd_17466_half_map_1.map.gz emd_17466_half_map_2.map.gz emd_17466_half_map_2.map.gz | 6 MB 6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17466 http://ftp.pdbj.org/pub/emdb/structures/EMD-17466 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17466 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17466 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_17466.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17466.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | DeepEMhancer map of UBR5~UbD~UbA | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.77 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half map of UBR5~UbD~UbA
| File | emd_17466_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map of UBR5~UbD~UbA | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map of UBR5~UbD~UbA
| File | emd_17466_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map of UBR5~UbD~UbA | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : HECT E3 UBR5 forming K48 linked Ubiquitin chains
| Entire | Name: HECT E3 UBR5 forming K48 linked Ubiquitin chains |
|---|---|
| Components |
|
-Supramolecule #1: HECT E3 UBR5 forming K48 linked Ubiquitin chains
| Supramolecule | Name: HECT E3 UBR5 forming K48 linked Ubiquitin chains / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Molecular weight | Theoretical: 620 KDa |
-Supramolecule #2: Ubiquitin
| Supramolecule | Name: Ubiquitin / type: complex / ID: 2 / Parent: 1 / Details: Donor Ubiquitin UbD |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 9 KDa |
-Supramolecule #3: Ubiquitin
| Supramolecule | Name: Ubiquitin / type: complex / ID: 3 / Parent: 2 / Details: Acceptor Ubiquitin UbA |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 9 KDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.3 µm |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)