+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Nucleocapsid protein Sars-Cov2 | |||||||||
 Map data Map data | map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Nucleocapsid protein / Sars-Cov2 / N protein / RNA BINDING PROTEIN | |||||||||
| Biological species |  | |||||||||
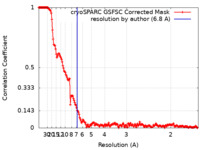
| Method | single particle reconstruction / cryo EM / Resolution: 6.8 Å | |||||||||
 Authors Authors | Farci D / Piano D / Graca AT | |||||||||
| Funding support |  Poland, 1 items Poland, 1 items
| |||||||||
 Citation Citation |  Journal: J Struct Biol / Year: 2025 Journal: J Struct Biol / Year: 2025Title: Characterization of SARS-CoV-2 nucleocapsid protein oligomers. Authors: Domenica Farci / André T Graça / Michael Hall / Patrycja Haniewicz / Sami Kereïche / Peter Faull / Joanna Kirkpatrick / Enzo Tramontano / Wolfgang P Schröder / Dario Piano /       Abstract: Oligomers of the SARS-CoV-2 nucleocapsid (N) protein are characterized by pronounced instability resulting in fast degradation. This property likely relates to two contrasting behaviors of the N ...Oligomers of the SARS-CoV-2 nucleocapsid (N) protein are characterized by pronounced instability resulting in fast degradation. This property likely relates to two contrasting behaviors of the N protein: genome stabilization through a compact nucleocapsid during cell evasion and genome release by nucleocapsid disassembling during infection. In vivo, the N protein forms rounded complexes of high molecular mass from its interaction with the viral genome. To study the N protein and understand its instability, we analyzed degradation profiles under different conditions by size-exclusion chromatography and characterized samples by mass spectrometry and cryo-electron microscopy. We identified self-cleavage properties of the N protein based on specific Proprotein convertases activities, with Cl playing a key role in modulating stability and degradation. These findings allowed isolation of a stable oligomeric complex of N, for which we report the 3D structure at ∼6.8 Å resolution. Findings are discussed considering available knowledge about the coronaviruses' infection cycle. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17430.map.gz emd_17430.map.gz | 246.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17430-v30.xml emd-17430-v30.xml emd-17430.xml emd-17430.xml | 15.6 KB 15.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_17430_fsc.xml emd_17430_fsc.xml | 17 KB | Display |  FSC data file FSC data file |
| Images |  emd_17430.png emd_17430.png | 34.2 KB | ||
| Masks |  emd_17430_msk_1.map emd_17430_msk_1.map | 512 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-17430.cif.gz emd-17430.cif.gz | 4.3 KB | ||
| Others |  emd_17430_half_map_1.map.gz emd_17430_half_map_1.map.gz emd_17430_half_map_2.map.gz emd_17430_half_map_2.map.gz | 474.6 MB 474.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17430 http://ftp.pdbj.org/pub/emdb/structures/EMD-17430 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17430 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17430 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_17430.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17430.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.82 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_17430_msk_1.map emd_17430_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
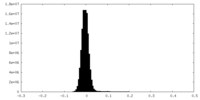
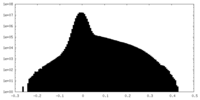
| Density Histograms |
-Half map: half map A
| File | emd_17430_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map B
| File | emd_17430_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Nucleocapsid protein Sars-Cov2
| Entire | Name: Nucleocapsid protein Sars-Cov2 |
|---|---|
| Components |
|
-Supramolecule #1: Nucleocapsid protein Sars-Cov2
| Supramolecule | Name: Nucleocapsid protein Sars-Cov2 / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 6 |
|---|---|
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 2 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: COUNTING / Digitization - Frames/image: 1-40 / Number grids imaged: 1 / Number real images: 9013 / Average electron dose: 59.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 165000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: AB INITIO MODEL |
|---|
 Movie
Movie Controller
Controller



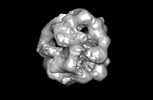

 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)