[English] 日本語
 Yorodumi
Yorodumi- EMDB-17385: Cryo-EM structure of the anaerobic ribonucleotide reductase from ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the anaerobic ribonucleotide reductase from Prevotella copri in its dimeric, dGTP/ATP-bound state | |||||||||||||||
 Map data Map data | Map post-processed with DeepEMhancer | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | ribonucleotide reductase / glycyl radical enzyme / allosteric regulation / nucleotide biosynthesis / OXIDOREDUCTASE / BIOSYNTHETIC PROTEIN | |||||||||||||||
| Function / homology | Ribonucleoside-triphosphate reductase, anaerobic / Anaerobic ribonucleoside-triphosphate reductase / ribonucleoside-triphosphate reductase (thioredoxin) activity / ATP-cone domain / ATP cone domain / ATP-cone domain profile. / DNA replication / ATP binding / Anaerobic ribonucleoside-triphosphate reductase Function and homology information Function and homology information | |||||||||||||||
| Biological species |  Prevotella copri (bacteria) / Prevotella copri (bacteria) /  Segatella copri (bacteria) Segatella copri (bacteria) | |||||||||||||||
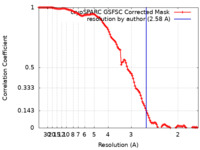
| Method | single particle reconstruction / cryo EM / Resolution: 2.58 Å | |||||||||||||||
 Authors Authors | Bimai O / Banerjee I / Sjoberg BM / Logan DT | |||||||||||||||
| Funding support |  Sweden, Sweden,  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Elife / Year: 2024 Journal: Elife / Year: 2024Title: Nucleotide binding to the ATP-cone in anaerobic ribonucleotide reductases allosterically regulates activity by modulating substrate binding. Authors: Ornella Bimai / Ipsita Banerjee / Inna Rozman Grinberg / Ping Huang / Lucas Hultgren / Simon Ekström / Daniel Lundin / Britt-Marie Sjöberg / Derek T Logan /  Abstract: A small, nucleotide-binding domain, the ATP-cone, is found at the N-terminus of most ribonucleotide reductase (RNR) catalytic subunits. By binding adenosine triphosphate (ATP) or deoxyadenosine ...A small, nucleotide-binding domain, the ATP-cone, is found at the N-terminus of most ribonucleotide reductase (RNR) catalytic subunits. By binding adenosine triphosphate (ATP) or deoxyadenosine triphosphate (dATP) it regulates the enzyme activity of all classes of RNR. Functional and structural work on aerobic RNRs has revealed a plethora of ways in which dATP inhibits activity by inducing oligomerisation and preventing a productive radical transfer from one subunit to the active site in the other. Anaerobic RNRs, on the other hand, store a stable glycyl radical next to the active site and the basis for their dATP-dependent inhibition is completely unknown. We present biochemical, biophysical, and structural information on the effects of ATP and dATP binding to the anaerobic RNR from . The enzyme exists in a dimer-tetramer equilibrium biased towards dimers when two ATP molecules are bound to the ATP-cone and tetramers when two dATP molecules are bound. In the presence of ATP, NrdD is active and has a fully ordered glycyl radical domain (GRD) in one monomer of the dimer. Binding of dATP to the ATP-cone results in loss of activity and increased dynamics of the GRD, such that it cannot be detected in the cryo-EM structures. The glycyl radical is formed even in the dATP-bound form, but the substrate does not bind. The structures implicate a complex network of interactions in activity regulation that involve the GRD more than 30 Å away from the dATP molecules, the allosteric substrate specificity site and a conserved but previously unseen flap over the active site. Taken together, the results suggest that dATP inhibition in anaerobic RNRs acts by increasing the flexibility of the flap and GRD, thereby preventing both substrate binding and radical mobilisation. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17385.map.gz emd_17385.map.gz | 143.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17385-v30.xml emd-17385-v30.xml emd-17385.xml emd-17385.xml | 25.7 KB 25.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_17385_fsc.xml emd_17385_fsc.xml | 11.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_17385.png emd_17385.png | 56 KB | ||
| Masks |  emd_17385_msk_1.map emd_17385_msk_1.map | 163.6 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-17385.cif.gz emd-17385.cif.gz | 7.4 KB | ||
| Others |  emd_17385_additional_1.map.gz emd_17385_additional_1.map.gz emd_17385_half_map_1.map.gz emd_17385_half_map_1.map.gz emd_17385_half_map_2.map.gz emd_17385_half_map_2.map.gz | 82.4 MB 151.9 MB 151.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17385 http://ftp.pdbj.org/pub/emdb/structures/EMD-17385 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17385 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17385 | HTTPS FTP |
-Related structure data
| Related structure data |  8p39MC  8p23C  8p27C  8p28C  8p2cC  8p2dC  8p2sC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_17385.map.gz / Format: CCP4 / Size: 163.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17385.map.gz / Format: CCP4 / Size: 163.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map post-processed with DeepEMhancer | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.86 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_17385_msk_1.map emd_17385_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Unsharpened map from non-uniform refinement with cryoSPARC
| File | emd_17385_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened map from non-uniform refinement with cryoSPARC | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map A from non-uniform refinement with cryoSPARC
| File | emd_17385_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A from non-uniform refinement with cryoSPARC | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map B from non-uniform refinement with cryoSPARC
| File | emd_17385_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B from non-uniform refinement with cryoSPARC | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Anaerobic ribonucleotide reductase from Prevotella copri in its d...
| Entire | Name: Anaerobic ribonucleotide reductase from Prevotella copri in its dimeric, dGTP/ATP-bound state |
|---|---|
| Components |
|
-Supramolecule #1: Anaerobic ribonucleotide reductase from Prevotella copri in its d...
| Supramolecule | Name: Anaerobic ribonucleotide reductase from Prevotella copri in its dimeric, dGTP/ATP-bound state type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Prevotella copri (bacteria) Prevotella copri (bacteria) |
| Molecular weight | Theoretical: 168.4 KDa |
-Macromolecule #1: Anaerobic ribonucleoside-triphosphate reductase
| Macromolecule | Name: Anaerobic ribonucleoside-triphosphate reductase / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Segatella copri (bacteria) Segatella copri (bacteria) |
| Molecular weight | Theoretical: 84.636055 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GPGSMIQTVV KRDGRIVGFN EQKIMAAIRK AMLHTDKGED TTLIEQITDH ISYRGKSQMS VEAIQDAIEM ELMKSARKDV AQKYIAYRN QRNIARKAKT RDVFMSIVNA KNNDITRENA NMNADTPAGM MMKFASETTK PFVDDYLLSE DVRDAVMHNY I HIHDKDYY ...String: GPGSMIQTVV KRDGRIVGFN EQKIMAAIRK AMLHTDKGED TTLIEQITDH ISYRGKSQMS VEAIQDAIEM ELMKSARKDV AQKYIAYRN QRNIARKAKT RDVFMSIVNA KNNDITRENA NMNADTPAGM MMKFASETTK PFVDDYLLSE DVRDAVMHNY I HIHDKDYY PTKSLTCVQH PLDVILNHGF TAGHGSSRPA KRIETAAVLA CISLETCQNE MHGGQAIPAF DFYLAPYVRM SY QEEVKNL EKLTGEDLSN LYDAPIDDYI EKPLDGLQGR ERLEQHAINK TVNRVHQAME AFIHNMNTIH SRGGNQVVFS SIN YGTDTS AEGRCIMREI LQSTYQGVGN GETAIFPIQI WKKKRGVNYL PEDRNYDLYK LACKVTARRF FPNFLNLDAT FNQN EKWRA DDPERYKWEI ATMGCRTRVF EDRWGEKTSI ARGNLSFSTI NIVKLAIECM GIENEKQRID MFFAKLDNIL DITAK QLDE RFQFQKTAMA KQFPLLMKYL WVGAENLKPE ETIESVINHG TLGIGFIGLA ECLVALIGKH HGESEKAQEL GLKIIT YMR DRANEFSEQY HHNYSILATP AEGLSGKFTK KDRKQFGVIP GVTDRDYYTN SNHVPVYYKC TALKKAQIEA PYHDLTR GG HIFYVEIDGD ATHNPSVIES VVDMMDKYNM GYGSVNHNRN RCLDCGYENA DAHLEVCPKC GSHHIDKLQR ITGYLVGT T DRWNSGKLAE LHDRVTHIGG EK UniProtKB: Anaerobic ribonucleoside-triphosphate reductase |
-Macromolecule #2: 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 2 / Number of copies: 2 / Formula: DGT |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-DGT: |
-Macromolecule #3: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 3 / Number of copies: 2 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #4: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 4 / Number of copies: 1 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Macromolecule #5: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 5 / Number of copies: 1 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||||||||||||||
| Grid | Model: Quantifoil R2/1 / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Details: GloQube, 20 mA | |||||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV / Details: blot force 1, 5s blot time. |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Number grids imaged: 1 / Number real images: 14346 / Average exposure time: 2.0 sec. / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.4 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | Chain - Source name: Other / Chain - Initial model type: experimental model Details: Half of a previously-built tetrameric form of the same enzyme |
|---|---|
| Details | Manual fitting was done using Coot and automatic real space refinement used phenix.refine |
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Target criteria: correlation coefficient |
| Output model |  PDB-8p39: |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)