+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human N-deacetylase/N-sulfotransferase 1 homodimer | |||||||||||||||
 Map data Map data | EM map of human NDST1 homodimer | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | heparan sulfate / bifunctional enzyme / Golgi / de-acetylatase & sulfotransferease / BIOSYNTHETIC PROTEIN | |||||||||||||||
| Function / homology |  Function and homology information Function and homology information[heparan sulfate]-glucosamine N-sulfotransferase / heparan sulfate N-sulfotransferase activity / heparan sulfate N-deacetylase activity / N-acetylglucosamine deacetylase activity / heparin proteoglycan biosynthetic process / embryonic neurocranium morphogenesis / embryonic viscerocranium morphogenesis / HS-GAG biosynthesis / cardiac septum development / glycosaminoglycan metabolic process ...[heparan sulfate]-glucosamine N-sulfotransferase / heparan sulfate N-sulfotransferase activity / heparan sulfate N-deacetylase activity / N-acetylglucosamine deacetylase activity / heparin proteoglycan biosynthetic process / embryonic neurocranium morphogenesis / embryonic viscerocranium morphogenesis / HS-GAG biosynthesis / cardiac septum development / glycosaminoglycan metabolic process / heparan sulfate proteoglycan biosynthetic process / deacetylase activity / respiratory gaseous exchange by respiratory system / polysaccharide biosynthetic process / coronary vasculature development / positive regulation of smoothened signaling pathway / Hydrolases; Acting on carbon-nitrogen bonds, other than peptide bonds; In linear amides / aorta development / midbrain development / forebrain development / fibroblast growth factor receptor signaling pathway / trans-Golgi network membrane / cell population proliferation / positive regulation of MAPK cascade / inflammatory response / Golgi membrane / Golgi apparatus Similarity search - Function | |||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.5 Å | |||||||||||||||
 Authors Authors | Vallet SD / Lortat-Jacob H / Wild R | |||||||||||||||
| Funding support |  France, 4 items France, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Proteoglycan Res / Year: 2023 Journal: Proteoglycan Res / Year: 2023Title: Functional and structural insights into human N-deacetylase/N-sulfotransferase activities Authors: Vallet SD / Annaval T / Vives RR / Richard E / Henault J / Le Narvor C / Bonnaffe D / Priem B / Wild R / Lortat-Jacob H | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17349.map.gz emd_17349.map.gz | 117.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17349-v30.xml emd-17349-v30.xml emd-17349.xml emd-17349.xml | 19.5 KB 19.5 KB | Display Display |  EMDB header EMDB header |
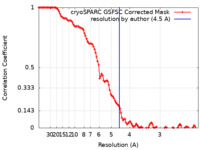
| FSC (resolution estimation) |  emd_17349_fsc.xml emd_17349_fsc.xml | 10.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_17349.png emd_17349.png | 106.1 KB | ||
| Masks |  emd_17349_msk_1.map emd_17349_msk_1.map | 125 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-17349.cif.gz emd-17349.cif.gz | 6.3 KB | ||
| Others |  emd_17349_half_map_1.map.gz emd_17349_half_map_1.map.gz emd_17349_half_map_2.map.gz emd_17349_half_map_2.map.gz | 116 MB 116 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17349 http://ftp.pdbj.org/pub/emdb/structures/EMD-17349 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17349 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17349 | HTTPS FTP |
-Related structure data
| Related structure data |  9f6zM M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_17349.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17349.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | EM map of human NDST1 homodimer | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.125 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_17349_msk_1.map emd_17349_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map B from final non-uniform refinement
| File | emd_17349_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B from final non-uniform refinement | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map A from final non-uniform refinement
| File | emd_17349_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A from final non-uniform refinement | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : NDST1 homodimer
| Entire | Name: NDST1 homodimer |
|---|---|
| Components |
|
-Supramolecule #1: NDST1 homodimer
| Supramolecule | Name: NDST1 homodimer / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 220 KDa |
-Macromolecule #1: N-deacetylase/N-sulfotransferase 1
| Macromolecule | Name: N-deacetylase/N-sulfotransferase 1 / type: protein_or_peptide / ID: 1 Details: Human NDST1 homodimer (residues 43-882), containing a linker and a 8-His tag at the C-terminus Enantiomer: LEVO / EC number: [heparan sulfate]-glucosamine N-sulfotransferase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GASRGLEPSA DAPEPDCGDP PPVAPSRLLP LKPVQAATPS RTDPLVLVFV ESLYSQLGQE VVAILESSR FKYRTEIAPG KGDMPTLTDK GRGRFALIIY ENILKYVNLD AWNRELLDKY C VAYGVGII GFFKANENSL LSAQLKGFPL FLHSNLGLKD CSINPKSPLL ...String: GASRGLEPSA DAPEPDCGDP PPVAPSRLLP LKPVQAATPS RTDPLVLVFV ESLYSQLGQE VVAILESSR FKYRTEIAPG KGDMPTLTDK GRGRFALIIY ENILKYVNLD AWNRELLDKY C VAYGVGII GFFKANENSL LSAQLKGFPL FLHSNLGLKD CSINPKSPLL YVTRPSEVEK GV LPGEDWT VFQSNHSTYE PVLLAKTRSS ESIPHLGADA GLHAALHATV VQDLGLHDGI QRV LFGNNL NFWLHKLVFV DAVAFLTGKR LSLPLDRYIL VDIDDIFVGK EGTRMKVEDV KALF DTQNE LRAHIPNFTF NLGYSGKFFH TGTNAEDAGD DLLLSYVKEF WWFPHMWSHM QPHLF HNQS VLAEQMALNK KFAVEHGIPT DMGYAVAPHH SGVYPVHVQL YEAWKQVWSI RVTSTE EYP HLKPARYRRG FIHNGIMVLP RQTCGLFTHT IFYNEYPGGS SELDKIINGG ELFLTVL LN PISIFMTHLS NYGNDRLGLY TFKHLVRFLH SWTNLRLQTL PPVQLAQKYF QIFSEEKD P LWQDPCEDKR HKDIWSKEKT CDRFPKLLII GPQKTGTTAL YLFLGMHPDL SSNYPSSET FEEIQFFNGH NYHKGIDWYM EFFPIPSNTT SDFYFEKSAN YFDSEVAPRR AAALLPKAKV LTILINPAD RAYSWYQHQR AHDDPVALKY TFHEVITAGS DASSKLRALQ NRCLVPGWYA T HIERWLSA YHANQILVLD GKLLRTEPAK VMDMVQKFLG VTNTIDYHKT LAFDPKKGFW CQ LLEGGKT KCLGKSKGRK YPEMDLDSRA FLKDYYRDHN IELSKLLYKM GQTLPTWLRE DLQ NTRNNN NNNGHHHHHH HH UniProtKB: Bifunctional heparan sulfate N-deacetylase/N-sulfotransferase 1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.3 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 6.5 Component:
| |||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 45 sec. / Pretreatment - Atmosphere: AIR / Details: Current: 25 mA | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV / Details: Blotted for 4 s with a blot force of 0. | |||||||||
| Details | Protein eluted as monodisperse peak from size exclusion chromatography column |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Frames/image: 1-44 / Number grids imaged: 1 / Number real images: 3038 / Average exposure time: 4.4 sec. / Average electron dose: 38.9 e/Å2 Details: Dataset was collected at a 30 degrees stage tilt angle |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 36000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | Chain - Residue range: 65-882 / Chain - Source name: AlphaFold / Chain - Initial model type: in silico model Details: Model of NDST1 homodimer was predicted using AlphaFold2 |
|---|---|
| Details | CCC 0.658 |
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Target criteria: cross-correlation coefficient |
| Output model |  PDB-9f6z: |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)